I've blogged before about how, for children under five, it's not the 'sexy' microbes that kill, but instead, the run of the mill ones: the bacteria that cause diarrhea and pneumonia are the culprits. One of the things I have heard a lot of recently regarding antibiotic development (and related therapies) is that we need to focus on 'non-paradigm' and non-model organisms. There's a problem with that approach:
The non-standard microbes aren't the ones causing the bulk of bacterial disease.
Oh dear.
In roaming around the International Society for Microbial Resistance website, I came across this telling figure:
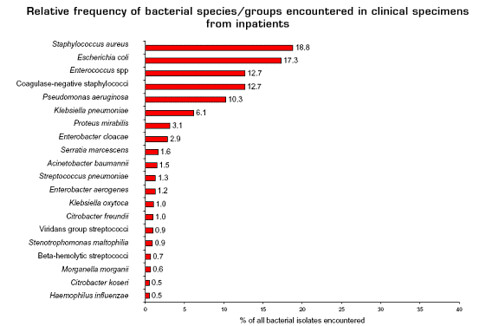
(from here)
This chart shows the frequencies of different species of bacteria that are isolated from sick people in hospitals--these include both hospital-acquired infections and infections that require hospitalization (a larger version can be found here). Many of these organisms might not be familiar, although, hopefully, Escherichia coli, Streptococcus, and Staphylococcus aureus (the methicillin resistant strains of Staphylococcus aureus are known as MRSA) are familiar.
What's important is that most of these bacteria are either 'model' organisms or close relatives of model organisms. For example, E. coli, perhaps the 'ur-model' organism, and its relatives (the Enterobacteriaceae) comprise 35.3% of the in-patient isolates. Let me make something clear. I'm not arguing that each of these species doesn't have its own ecology and set of pathologies: different groups of E. coli can differ radically in their ecologies and pathologies. But from a drug development perspective, if you're trying to identify novel drug targets they are very similar. As a professor of mine said, "Once you throw 'em in the blender, they pretty much look the same." (He was a wee bit reductionist).
The rest of the organisms, Enterococcus, the staphylococci, and streptococci are very well characterized, and are essentially Gram-positive model organisms. Pseudomonas aeruginosa is the non-enteric Gram-negative model organism. While Acinetobacter baumannii is increasing in frequency, and can have devastating effects [links], it hardly a new organism: the type species of Acinetobacter was identified in 1911 (like most bacterial taxonomy, Acinetobacter taxonomy is a disaster). The only thing that's even remotely an oddball is Haemophilus influenzae. Considering that H. influenzae was one the first genomes sequenced, it's hardly a bizarre organism.
When one looks at out-patient infections, the picture is virtually identical, although the rankings change slightly:
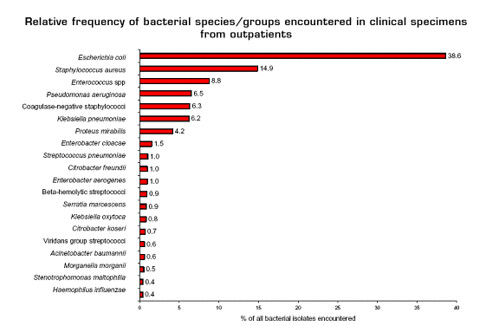
(from here; larger image here)
Notice that there are no spirochaetes on the list, and the really funky things like methanotroph anaerobes, well, let's just say that the medically important are a very restricted depauperate set of the existing microbial diversity. Rather than trying to target 'novel' outliers for drug development, we need to focus on new sources of antibiotics.
Note: I'm not saying that we shouldn't study oddball organisms for their own sake, but as a medical drug development strategy, it doesn't make much sense to me.
- Log in to post comments

Perhaps this is just my ignorance talking, but it seems to me to be a bit of a leap from isolating a bacterial species to identifying it as the cause of an infection.
Mike--Following up on Andrew's comment, do the stats for E. coli only refer to pathogenic strains? After all, we carry lots of harmless strains which will breed like crazy in a culture, so the fact that they dominate these surveys might not mean all that much.
These are the strains identified as the etiological agent. While some infections are polymicrobial (pneumonia) so it's hard to identify the causative organism in every case (although after decades of medical microbiology, we have a good idea), most of these organisms are isolated from typically sterile sites (urinary tracts, bloodstream), or from things like boils. I should have been clearer, but these are the etiological agents, and they are associated with pathologies. That's why I referred to them as infections, not commensal bacteria.
Ah, it was my ignorance talking. Thanks for the clarification.
Hi, guys. The chart on outpatient isolates might have a pretty significant source of bias. It seems like most of the data probably comes from urine cultures (E coli, Klebsiella, Proteus) and skin/wound cultures (S aureus, coag-neg Staph), since those are easy and common culture sites in the outpt setting. I do agree that treatment failure and pen or fluoroquinolone resistance are growing problems with E coli UTI (some studies: 40% amox-resistant, 20% FQ-resistant). But, for a lot of other bacterial illnesses, pathogen mix is changing, H flu and atypicals are on the rise, and treatment failures are big issues, though routine culture is not part of the standard outpt workup.
1. Acute otitis media:
Most clinics (ours included) use a treatment protocol based on knowledge of local pathogens and resistance patterns from studies using tympanocentesis (aspiration of fluid from the middle ear). Doing tympanocentesis routinely for every case carries a risk of complications and, as far as I know, does not improve outcomes. Thus, there's culture data only from the studies, not from the large number of cases treated without cx. Here's some approximate data on the pathogen distribution:
H flu about 60% (of which about half now beta-lactamase+)
S pneumo about 30% (declining due to vaccination in kids)
Moraxella catarrhalis about 10%
some Group A Strep (pyogenes)
2. Community-acquired pneumonia:
Again, as for AOM, most treatment is empiric, based on study data. Sputum cultures can be hard to get and yield a lot of bad data (contamination with mouth flora etc). Even blood cx in hospitalized CAP pts with severe diseases is usually negative. Approx pathogen distrib:
S pneumo 85%, also declining due to vaccination
Atypicals 15% (incl Mycoplasma spp, Chlamydia spp, Legionella)
Of note in CAP, atypicals are also on the rise, and they contribute significantly to overall morbidity/mortality and treatment cost, because they resist empiric tx and cause extrapulmonary disease out of proportion to their 15% frequency.
3. Bacterial tonsillitis/pharyngitis:
Group A Strep is by far the most common pathogen, but a lot of clinics just use the rapid Strep EIA rather than culture (our practice varies; most of us usu do both). So, for all the cases with only EIA, no cx data. Of note, some "non-boring" atypicals (including Mycoplasma) are make up increasing numbers of cases.
So, to get a more accurate distribution, it seems that we would need to correct for bacterial illnesses that don't routinely get cultured, and, if we're talking about logical driving forces for new antimicrobial development, correct again to reflect pathogens that are more likely to cause treatment failures and more severe disease.
Also, we could benefit from good adherence to a sound empiric treatment protocol for UTI based on up-to-date E coli data. Even with routine culture, symptomatic pts will generally be started on abx before a final report (with sensitivities)comes back.
Sorry I'm coming late to this discussion. I followed the link from a recent post on E coli commensals.
What are you angry about?
how can you cope with microbes what are microbes? i need to know for my project because i am in discovery at a secondry school.
how can you cope with microbes what are microbes? i need to know for my project because i am in discovery at a secondry school.