The image below is a phylogram, illustrating the degree of variation in a sequence of mitochondrial DNA. The concept is fairly simple: if two DNA samples are from individuals that are evolutionarily distant from one another, they'll have accumulated more differences in their mitochondrial DNA, and will be drawn farther apart from one another. If the two individuals are closely related, their DNA will be more similar, and they'll be drawn closer together. That's the key thing you need to know to understand what's going on.
There are other, more complicated analyses going on in the figure, too: the branching pattern is determined by analyzing subsets of shared sequences, and it takes a fair bit of computing power to put the full picture together. You'll just have to trust me on that one, but all you need to know is that the branches are objectively calculated, and that the distances between the tips of the branches and their last branching point tell you something about the degree of genetic disparity in the group.
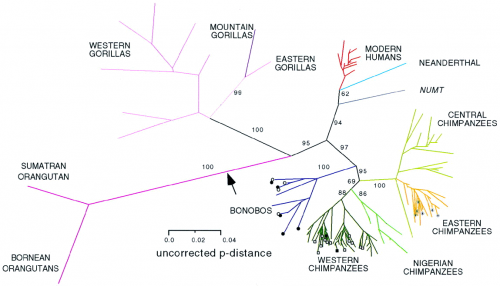 Unrooted phylogram of mitochondrial DNA sequences.
Unrooted phylogram of mitochondrial DNA sequences.
Gagneux P1, Wills C, Gerloff U, Tautz D, Morin PA, Boesch C, Fruth B, Hohmann G, Ryder OA, Woodruff DS. (1999) Mitochondrial sequences show diverse evolutionary histories of African hominoids. Proc Natl Acad Sci USA 96(9):5077-82.
Isn't that a lovely family tree? You can see that the orangutan branch is a long, long way from the human branch, as expected, and that the chimpanzee and human branch point is closer to us than to the point where gorillas diverge from chimpanzees and human, also as expected.
But look at the modern humans, in red. We're a sadly cramped little cluster; there may be a lot of us individuals, but we're all relatively closely related. Modern humans only arose roughly 100,000 years ago, and our branch expanded numerically fairly rapidly, but we haven't had all that much time to accumulate as much genetic variation. We're a close-knit side branch of the primate family tree, or we ought to be close-knit -- we're brothers and sisters together.
We're definitely set somewhat apart from Cousin Neanderthal, the light blue line. But even there, the range of genetic diversity is pitiful compared to the chimpanzees.
The exuberant flowering of the chimpanzee lineage, all those blue and green and orange lines, tells us that these come from deep, distinct populations. All those beings we just lump together as chimps are actually genetically diverse, representing multiple long-established lineages -- Central Chimpanzees are more different from Western Chimpanzees than we are from Neanderthal. Western Gorillas are far more distant from Eastern Gorillas than any branch of the modern human tree. The orangutans of Sumatra and Borneo, two islands separated by a few hundred kilometers, show far greater genetic differences than Chinese and African humans.
The patterns of descent we see in that stunted bud of humanity may be real, but they're tiny when compared to the grand bouquet of chimpanzee genetics. So how can we fight over the few small superficial differences that mark human races? I guess we're just really good at squabbling with our brothers and sisters.
- Log in to post comments

The term race implies greater differences than exist, I think variety might be a more accurate word. Not holding my breath waiting for it to catch on...
BTW, numt means "nuclear copy of the mitochondrial genome". Such things arise when a mitochondrion is destroyed and its genome ends up in the nucleus (perhaps after mitosis, during which the nucleus doesn't exist and all the DNA is set free).
Has anyone heard of the new CLIA Grade products - http://www.americanbio.com - seems to be a great choice for molecular diagnostics and more.
i don't see the new world primates on this genetic tree. Too far removed to mark the cut?
Tim H.: "Variety" and "Race" traditionally mean the same thing as each other, and as "subspecies" Variety is used for plants. This is all arcane, but we're stuck with those prior uses.
"geographic ancestry" should replace it all. great post. thanks.
What exactly is this article after? Some anti-racialist rhetoric? The diversity of lower primate genetics is easily explained by the fact that humans are exclusive in their ability to adapt their habitat rather than their genetics in order to thrive, which then precludes the necessity of wildly diverse genetic populations.
On top of which is the amble evidence that the various races have peculiar traits such as differing blood factors, skeletal densities, and so on. Bi-racial individuals have been noted to be at much higher risk of medical complications from birth, and tri-racial even more. The most evident conclusion is that human races are different enough to be scientifically relevant, and it's patently unscientific to claim race is superficial. It's like saying breeds of dogs are superficial and you don't understand why your mutt has severe hip dysplasia because of it.
"What exactly is this article after? Some anti-racialist rhetoric?"
Uh, Nick, you say that as if it were a bad thing
Way too far removed to be included in this picture! You'll notice even the Old World monkeys aren't included, and they're closer.
O RLY?
[citation needed]
Not convinced Nick is on this tree, either.
Of course our “sadly cramped little cluster” isn't helped with Neanderthal being a separate species.
Domestic animal races can be generated in a short time scale. It is not surprising that large visible differences are not correlated to ancient divergence.
I am confused on "uncorrected p-distance" For example the very long line from the point separating Orangutans from Gorillas is labeled "100". Another line leading from this point towards Gorillas is also labeled "100". My problem is that these two lines are very different in length. I could cite many other examples along similar lines. What am I missing?
The numbers on the branches are statistical support for the grouping of sequences within each clade. So 100 means that 100% of analyses found the result for a particular group. P-distance refers to percentage difference, as this network analysis is based on a genetic distance calculation (phenetic) rather than shared derived traits (cladistic).
And the "uncorrected" just means that no model of nucleotide evolution was used (e.g., K2P or GTR). This is the raw data as visualized most simply--pretty cool to see the amount of genetic diversity even within chimps. I wonder, being that this study is 15 years old and based on mtDNA, what are the more recent genome-wide studies showing?
The values for p-distance are defined by the branch lengths. The numerical labels affectively refer (I think) to a percentage confidence from the analysis of all the members linked* to that branch belonging together in a natural group, i.e. form a clade.
*from the "outside" end! Difficult to read in an unrooted tree.