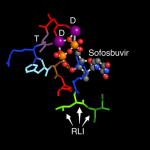
BLAST
It's well understood in science education that students are more engaged when they work on problems that matter. Right now, Zika virus matters. Zika is a very scary problem that matters a great deal to anyone who might want to start a family and greatly concerns my students.
I teach a bioinformatics course where students use computational tools to research biology. Since my students are learning how to use tools that can be applied to this problem, I decided to have them apply their new bioinformatics skills to identify drugs that work against Zika virus.
We don't have the lab facilities…
Sometimes when you go digging through the databases, you find unexpected things.
When I was researching the previous posts on insulin structure and insulin evolution, I found something curious indeed.
Human insulin, colored by rainbow. Image from the Molecule World iPad app by Digital World Biology.
I wanted to find out how many different organisms made insulin, so I used a database at the NCBI called Blink. Blink is a database of protein blast search results. Using Blink can save you lots of time because it organizes blast results from all the organisms in the non-…
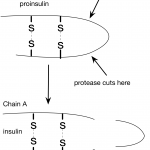
In my last post, I wrote about insulin and interesting features of the insulin structure. Some of the things I learned were really surprising. For example, I was surprised to learn how similar pig and human insulin are. I hadn't considered this before, but this made me wonder about the human insulin we used to give to one of our cats. How do cat and human insulin compare?
It turns out, that all vertebrates produce insulin, even frogs and zebra fish. Human preproinsulin is only 110 amino acids long and even human and fish insulin are pretty similar. Of course, this observation only leads…
Thanks to the internet, you can find out your pirate name and your Jersey Shore name, and now thanks to the EMBL-EBI learning tools, you can find your protein name too!
When you type your name into the box, the program reads the letters of your name as if they were the single-letter codes for amino acids. Since there are only 20 amino acids, if you have a B, J, O, U, X, or Z in your name the program reads it as "X" which just means any amino acid could go in that spot.
The amino acids are then translated back into one of the possible three-letter DNA codes for each amino acid, and that DNA…
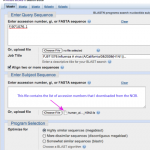
No more delays! BLAST away!
Time to blast. Let's see what it means for sequences to be similar.
First, we'll plan our experiment. When I think about digital biology experiments, I organize the steps in the following way:
A. Defining the question
B. Making the data sets
C. Analyzing the data sets
D. Interpreting the results
I'm going intersperse my results with a few instructions so you can repeat the things that I've done below. I've some people writing that only experts should be analyzing data. But I disagree with those who say that sequence…
We'll have a blast, I promise! But there's one little thing we need to discuss first...
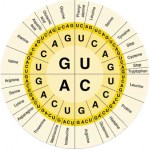
I want to explain why I'm going to use nucleotide sequences for the blast search. (I used protein the other day). It's not just because someone told me too, there is a solid rational reason for this.
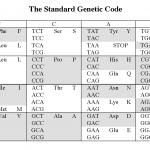
The reason is the redundancy in the genetic code.
Okay, that probably didn't make any sense to those of you who didn't already know the answer. Here it is.
The picture above shows the human genetic code (there are at least 16 variations on this, but that's another story). Each middle cell in the table…
In which we identify unknown human proteins.
Yesterday, I wrote about using the BLOSUM 62 matrix to calculate a score for matches between two proteins. Those scores give us a good start on understanding how blastp determines whether two sequences are matching by chance or because they're more likely to be related. But that's not all there is to calculating a blast score, and there's at least one other statistic to consider as well, the E value.
It all comes down to biochemistry
The BLOSUM 62 matrix is based on the substitutions that really do or do not happen in real protein sequences. I…
In which we search for Elvis, using blastp, and find out how old we would have to be to see Elvis in a Las Vegas club.
Introduction Once you're acquainted with proteins, amino acids, and the kinds of bonds that hold proteins together, we can talk about using this information to evaluate the similarity between protein sequences. We can easily imagine that if two protein sequences are identical, then those proteins would have the same kind of activity. But what about proteins that are similar in some regions, and not others, or proteins that only share some of the same amino acids in similar…
Ebola virus has impressed me as creepy ever since I read "The Hot Zone: A Terrifying True Story some years back by Richard Preston. (I guess he has a new book, too, Panic in Level 4: Cannibals, Killer Viruses, and Other Journeys to the Edge of Science but I haven't been in airport for the past couple of weeks, so I haven't read it yet.)
Technorati Tags: blast, phylogenetic trees, Ebola, viruses
Infectious agents that cause diseases with gruesome symptoms really excite those of us with an interest in microbiology. Tara has written about this paper, too, and summarized the details.
I…
Let's play anomaly!
Most of this week, I've written about the fun time I had playing around with NCBI's Blink database and finding evidence that at least one mosquito, Aedes aegypti, seems to have been infected at some point with a plant paramyxovirus and that the paramyxovirus left one of its genes behind, stuck in the mosquito genome.
During this process, I realized that the method I used works with other viruses, too. I tried it with a few random viruses and sure enough, I found some interesting things.
You've got a week to give it a try. Let's see what you find! The method is…
Do mosquitoes get the mumps? Part V. A general method for finding interesting things in GenBank
This is the last in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes and a general method for finding other interesting things.
In this last part, I discuss a general method for finding novel things in GenBank and how this kind of project could be a good sort of discovery, inquiry-based project for biology, microbiology, or bioinformatics students.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III.…
Part IV. Assembling the details and making the case for a novel paramyxovirus
This is the fourth in a five part series on an unexpected discovery of a paramyxovirus in a mosquito. In this part, we take a look at all the evidence we can find and try to figure out how a gene from a virus came to be part of the Aedes aegypti genome.
image from the Public Health Library
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a novel mosquito paramyxovirus
V. A…
Part III. Serendipity strikes when we Blink
In which we find an unexpected result when we Blink while looking at the mumps polymerase.
This is the third in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes. And yes, this is where the discovery happens.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a mosquito paramyxovirus
V. A general method for finding interesting things in GenBank
To paraphrase Louis Pasteur,…
Part II. What do mumps proteins do? And how do we find out?
This is the second in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes, and a general method for finding interesting things.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a mosquito paramyxovirus
V. A general method for finding interesting things in GenBank
In Part I, we looked at the NCBI SeqViewer, and found a new way to check out a genome map, and learn more…
Part I. The back story from the genome record
Together, these five posts describe the discovery of a novel paramyxovirus in the Aedes aegyptii genome and a new method for finding interesting anomalies in GenBank.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a mosquito paramyxovirus
V. A general method for finding interesting things in GenBank
I began this series on mumps intending to write about immunology and how vaccines work to stimulate the immune…
yep, I've become a videoblogger, at least sometimes.
See the first video below. Be kind in the comments, this is a new thing for me.
This video introduces the different blast programs, discusses word size, and how blastn works, the blastn score and the E value. The treatment is light and not too in depth, but as I said, it's an introduction.
A quick introduction to BLAST from Sandra Porter on Vimeo.
In which we're reminded that database searches are experiments, too.
One of the trickiest things with bioinformatics experiments is repeating them. This challenge isn't related to the validity of the original results, the challenge is that, unless you made your own database and kept it in the same state, the database that you'll be using at a later time, sometimes even a day later, is a different database. And, if you query a different database, you may get a different result.
The series that I'm currently posting is one that I started working on a couple of years ago. Originally, I was…
Last year I wrote about an experiment where I compared a human mitochondrial DNA sequence to primate sequences in the GenBank. Since I wanted to know about the differences between humans, gorillas, and chimps, I used the Entrez query 'Great Apes' to limit my search to a set of sequences in the PopSet database that contained gorillas, bonobos, chimps, and human DNA.
A week ago, I tried to repeat this experiment and...
It didn't work.
All I saw were human mitochondrial sequences. I know the other sequences match, but I didn't see them since there are so many human sequences that match…
We have lots of DNA samples from bacteria that were isolated from dirt. Now it's time to our own metagenomics project and figure out what they are. Our class project is on a much smaller scale than the honeybee metagenomics project that I wrote about yesterday, but we're using many of the same principles.
The general process is this:
1. We sort the chromatogram data to identify good data and separate it from bad data. Informatics can help you determine if data is good, and measure how good it is, but it cannot turn bad data into good data. And, there's no point in wasting time with…