NCBI
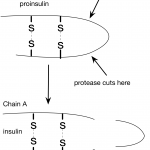
In my last post, I wrote about insulin and interesting features of the insulin structure. Some of the things I learned were really surprising. For example, I was surprised to learn how similar pig and human insulin are. I hadn't considered this before, but this made me wonder about the human insulin we used to give to one of our cats. How do cat and human insulin compare?
It turns out, that all vertebrates produce insulin, even frogs and zebra fish. Human preproinsulin is only 110 amino acids long and even human and fish insulin are pretty similar…
You might think the coolest thing about the Next Generation DNA Sequencing technologies is that we can use them to sequence long-dead mammoths, entire populations of microbes, or bits of bone from Neanderthals.
But you would be wrong.
Sure, those are all cool things to do, but Next Generation DNA sequencing (or NGS for short) can give us answers to questions that are far, far more interesting.
With NGS, we can look at entire transcriptomes (!!) together with the proteins that make them and the DNA modifications that help regulate them. If we compare a cell to music, a genome sequence…
No more delays! BLAST away!
Time to blast. Let's see what it means for sequences to be similar.
First, we'll plan our experiment. When I think about digital biology experiments, I organize the steps in the following way:
A. Defining the question
B. Making the data sets
C. Analyzing the data sets
D. Interpreting the results
I'm going intersperse my results with a few instructions so you can repeat the things that I've done…
We'll have a blast, I promise! But there's one little thing we need to discuss first...
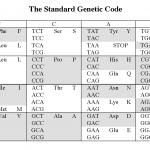
I want to explain why I'm going to use nucleotide sequences for the blast search. (I used protein the other day). It's not just because someone told me too, there is a solid rational reason for this.
The reason is the redundancy in the genetic code.
Okay, that probably didn't make any sense to those of you who didn't already know the answer. Here it is.
The picture above shows the human genetic code (there are at least 16 variations on this, but that's another story). Each middle cell in the…
In which we search for Elvis, using blastp, and find out how old we would have to be to see Elvis in a Las Vegas club.
Introduction
Once you're acquainted with proteins, amino acids, and the kinds of bonds that hold proteins together, we can talk about using this information to evaluate the similarity between protein sequences. We can easily imagine that if two protein sequences are identical, then those proteins would have the same kind of activity. But what about proteins that are similar in some regions, and not others, or proteins that only share some of the same amino acids in similar…
Have you ever wondered how to find things in the NCBI databases? Maybe you tried to find something but didn't know how it was spelled. Or maybe you tried to use a common name like "pig" or "deer" to find information in a database, not knowing that all the organism names are in Latin. Or perhaps you're wondering just what kind of information is stored for different kinds of records and if you could search for this information.
I wrote a book that covered this topic quite thoroughly, a couple of years ago, for the NCBI structure database. Now, I've decided to make some movies, too.
This…
Do mosquitoes get the mumps? Part V. A general method for finding interesting things in GenBank
This is the last in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes and a general method for finding other interesting things.
In this last part, I discuss a general method for finding novel things in GenBank and how this kind of project could be a good sort of discovery, inquiry-based project for biology, microbiology, or bioinformatics students.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III.…
Part IV. Assembling the details and making the case for a novel paramyxovirus
This is the fourth in a five part series on an unexpected discovery of a paramyxovirus in a mosquito. In this part, we take a look at all the evidence we can find and try to figure out how a gene from a virus came to be part of the Aedes aegypti genome.
image from the Public Health Library
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a novel mosquito paramyxovirus
V. A…
Part III. Serendipity strikes when we Blink
In which we find an unexpected result when we Blink while looking at the mumps polymerase.
This is the third in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes. And yes, this is where the discovery happens.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a mosquito paramyxovirus
V. A general method for finding interesting things in GenBank
To paraphrase Louis Pasteur,…
Part II. What do mumps proteins do? And how do we find out?
This is the second in a five part series on an unexpected discovery of a paramyxovirus in mosquitoes, and a general method for finding interesting things.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a mosquito paramyxovirus
V. A general method for finding interesting things in GenBank
In Part I, we looked at the NCBI SeqViewer, and found a new way to check out a genome map, and learn more…
Part I. The back story from the genome record
Together, these five posts describe the discovery of a novel paramyxovirus in the Aedes aegyptii genome and a new method for finding interesting anomalies in GenBank.
I. The back story from the genome record
II. What do the mumps proteins do? And how do we find out?
III. Serendipity strikes when we Blink.
IV. Assembling the details of the case for a mosquito paramyxovirus
V. A general method for finding interesting things in GenBank
I began this series on mumps intending to write about immunology and how vaccines work to stimulate the immune…
Instead of enjoying a sunny summer day today, or partying with SciBlings in New York, I'm staring out my window watching the rain. Inspiration hit! What about searching for August?
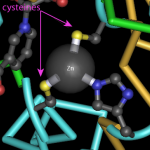
Folks, meet the HFQ protein from E. coli. I found this lovely molecule by doing a multi-database search at the NCBI with the term 'August'.
HFQ is a lovely protein with six identical subunits, that's involved in processing small RNA molecules and is homologous to some eucaryotic proteins that work in RNA splicing (1).
Do you see the blue loopy regions in the center of the structure? Those are positively…
or is it just an idea that's ahead of the curve?
Last week, I was stunned to discover at least 31 papers in an NCBI Gene database entry that were in the entry for the wrong gene. I wrote about this here, here, here, and here.
Now, an oversight like this is a little understandable. The titles of the entries do include the name of the wrong gene (DRD2 - the dopamine D2 receptor). And it was only four years ago that people figured out that the marker in the title of the articles mapped somewhere else.
If computers were responsible for the annotation, well, this would be understandable.…
It's pretty common these days to pick up an issue of Science or Nature and see people ranting about GenBank (1). Many of the rants are triggered, at least in part, by a wide-spread misunderstanding of what GenBank is and how it works. Perhaps this can be solved through education, but I don't think that's likely. People from the NCBI can explain over and over again that some of the sequence databases in GenBank are meant to be an archival resource (2), and define the term "archive," but that's not going to help.
Confusion about database content and oversight is widespread in this…
In a recent post, I wrote about an article that I read in Science magazine on the genetics of learning.
One of things about the article that surprised me quite a bit was a mistake the authors made in placing the polymorphism in the wrong gene. I wrote about that yesterday. The other thing that surprised me was something that I found at the NCBI.
The article that I wrote about definitely made a mistake and I don't understand why it wasn't caught by the reviewers. I found it pretty quickly by searching OMIM and I was only trying to find information about dopamine, not verify results.…