These days, DNA sequencing happens in one of three ways.
In the early days of DNA sequencing (like the 80's), labs prepared their own samples, sequenced those samples, and analyzed their results. Some labs still do this.
Then, in the 90's, genome centers came along. Genome centers are like giant factories that manufacture sequence data. They have buildings, dedicated staff, and professional bioinformaticians who write programs and work with other factory members to get the data entered, analyzed, and shipped out to the databases. (You can learn more about this and go on a virtual tour in this…
Next Saturday afternoon, at ScienceOnline2010, the science goddess, the chemspider, and I will be presenting a workshop on getting students involved in citizen science.
In preparation, I'm compiling a set of links to projects that involve students in citizen science. If you know of any good citizen science efforts, please share them in the comments.
Here we go!
Before I start listing links, I am limiting this list to projects that allow both students and citizen scientists to participate. I know of plenty of student projects, where students can isolate phage and annotate their genomes or…
This month's cover of The Scientist has a mistake that makes me cringe.
Can you spot what's wrong?
And they call themselves "The Scientist" humph!
Blaine Bettinger has an absolutely wonderful post where he compares his results for type 2 diabetes from 23andMe and DeCODEme.
I really liked his post and I appreciated the way he showed the data from the two companies and elaborated on their interpretation of his genotype and his risk.
Interestingly, his story goes beyond a simple relationship, where one base changes, one amino acid changes, and voila! you've got the disease.
Bettinger describes what happens when there are changes in multiple genes and how those changes can have a cumulative effect on evaluating the risk of developing a…
"What's in a name? that which we call a rose
By any other name would smell as sweet"
- Juliet, from Romeo and Juliet by William Shakespeare
I realized from the comments on my previous post and from Mike's post that more explanations were in order.
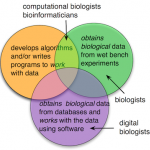
It seems we have two topics - why do we need a new name at all? and why the current names (biologist, computational biologist, bioinformatician, etc.) don't work. What really distinguishes a digital biologist from a regular, garden variety biologist? Why isn't a digital biologist a computational biologist?
So, I brought along two "show and tell…
If you're in Seattle, Dr. Bruce Alberts will be talking tomorrow night (Jan 5th) at the Seattle Aquarium on science education and the role that scientists play.
There are also some really interesting talks at a day-long workshop, Wednesday (Jan 6th) at the UW South Campus Center.
The details and registration info are below:
~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~
Tuesday:
COSEE Ocean Learning Communities & Washington SeaGrant Present
Redefining Science Education and the
Roles that Scientists Play in Society
Dr. Bruce Alberts
Tuesday,…
An NSF post on Twitter this morning described an interesting study from the University of Pennsylanvia and Cornell University, that found that some people who call themselves "African Americans" may only be 1% West African, according to their DNA.
The University of Pennsylvania press release contains other interesting findings as well. 365 individuals were studied and 300,000 genetic markers were examined.
Some of the findings were:
If you're African American, the genes most likely to have an African origin are those on your X chromosome. The article didn't mention it, but I would guess…
What do you call a biologist who uses bioinformatics tools to do research, but doesn't program?
You don't know?
Neither does anyone else.
The names we use
People who practice biology are known by many names, so many, that the number of names almost reflects the diversity of biology itself.
Sometimes we describe biologists by the subject they study. Thus, we have biologists from anatomists to zoologists, and everything in between: addiction researchers, chronobiologists, epidemiologists, immunologists, microbiologists, neuroscientists, pharmacologists, physiologists, plant biologists,…
I often get questions about bioinformatics, bioinformatics jobs and career paths. Most of the questions reflect a general sense of confusion between creating bioinformatics resources and using them. Bioinformatics is unique in this sense. No one confuses writing a package like Photoshop with being a photographer, yet for some odd reason, people seem to expect this of biologists. In the same respect, even the programmers and database administrators who work in bioinformatics, are unfairly assumed to have had graduate level training in biology.
In many ways, it's easiest to understand…
For many years, I've been perfectly content to work with small numbers of things. Working with one gene or one protein is great. Even small groups of genes are okay. I'm fine with alternatively spliced genes with multiple transcripts, or multiple polymorphisms, or genes in multi-gene families, or groups of genes in operons.
But I never trusted microarrays.
First, there were all the articles questioning the ability to reproduce microarray data. For many years, people reported difficulties in reproducing experiments, especially if they used chips from different manufacturers or different…
If you have a little time, the Dolan DNA Learning Center at Cold Spring Harbor will be presenting some really interesting workshops on neuroscience and genetics.
The dates are:
Nov. 5, 2009: Inside Cancer - workshop on teaching cancer; Raritan
Valley Community College, Somerville, NJ
Nov. 6, 2009: Genes To Cognition - workshop on teaching neuroscience;
Raritan Valley Community College, Somerville, NJ
Nov. 21, 2009: Inside Cancer - workshop on teaching cancer; Great Bay
Community College, Portsmouth, NH
Nov. 20, 2009: Genes To Cognition - workshop on teaching neuroscience;
Great Bay…
We always see interesting creatures whenever we walk on the beach. Now, a new program from the University of Washington and the state department of Fish and Wildlife is seeking to enlist beach walkers in a community science project where they can help monitor biodiversity.
The program is called NatureMapping and it's mission is to enlist the community and schools in monitoring the health of our beaches and contributing data to a state-managed biodiversity database. If you can get to a beach, you can help survey invertebrate species and intertidal marine fish.
Two free facilitator…
This morning I attended the Fifth Annual WBBA Governor's Life Sciences Summit. The breakfast was great; the talks were okay.
I do enjoy the stories about people who's lives were saved because of biotechnology and I agree that the focus of the summit, research and discovery are important, but I can't help thinking about the missing piece.
For the past ten years, I've been involved in a national experiment to help build an educated workforce for biotechnology. Through that time, I've learned about one glaring area where Washington state is missing out.
There's one word for the missing piece.…
How did it get between the windows?
Maybe this is why the flight attendant kept asking us to close the shades.
This summer, I had the good fortune to attend three (or was it four?) conferences on science education. One of the most inspirational conferences was one on Vision and Change in Biology Education. This conference was co-sponsored by the National Science Foundation and the AAAS. It was a call to action for biology educators and many of the points and findings resonated deep in my bones.
Then, I read the press release from the AAAS.
And right there in the middle, I found this statement from the AAAS CEO, Alan Leshner.
Leshner said the goal of undergraduate education should be to give students…
You might think the coolest thing about the Next Generation DNA Sequencing technologies is that we can use them to sequence long-dead mammoths, entire populations of microbes, or bits of bone from Neanderthals.
But you would be wrong.
Sure, those are all cool things to do, but Next Generation DNA sequencing (or NGS for short) can give us answers to questions that are far, far more interesting.
With NGS, we can look at entire transcriptomes (!!) together with the proteins that make them and the DNA modifications that help regulate them. If we compare a cell to music, a genome sequence…
I've just returned from two conferences that focused on educating students for careers in science and technology and what do I find here at the home fort? There's Chad writing a very nice series on science careers!
I was a little puzzled by PNAS acryonym in his titles since to me, PNAS stands for "Proceedings of the National Academy of Scientists" and is a high impact scientific journal. But then I realized that Chad is a physicist and he might not know this. It's quite possible that PNAS isn't as big in the physics community as it is in biology.
Anyway, this is a very nice series, so way to…
Last spring, I gave my first hands-on workshop in working with Next Generation Sequencing data at the Eighth Annual UT-ORNL-KBRIN Bioinformatics Summit at Fall Creek Falls State Park in Tennessee. The proceedings from that conference are now on-line at BMC Bioinformatics and it's fun to look back and reflect on all that I learned at the conference and all that's happened since.
Figure 1. Fall Creek Falls State Park, TN
When the conference took place, Geospiza had only just released new versions of GeneSifter Analysis Edition that could do gene expression analysis with Next Gen data.
Who…
I don't remember learning about plasma when I took physics, but it's amazing stuff. Last week at the Hi-Tec conference in Arizona, I got to learn how an electromagnetic field can be used to push plasma around a tube. Community college students get to play with the coolest toys!
Here's some plasma contained in a small area.
Figure 1. Plasma on the table.
Here's some plasma getting pushed around a tube.
Figure 2. Plasma getting pushed around.
Wikipedia has a very nice article on plasma and plasma displays that occur in nature, such as St. Elmo's fire. I guess if you want hands-on…
Liveblogging from the Hi-Tec conference
I'm currently at the Hi-Tec conference in Scottsdale, Arizona. (If you follow me on Twitter - www.twitter.com/@digitalbio - you may have seen me complaining about the temperature). It's an interesting conference, so I'm going to share some of the things that I'm learning.
Dr. Travis Benanti and Dr. Steve Fonash from Penn State University are presenting an interesting session this morning on nanotechnology.
Luckily, you don't have to know anything about nanotechnology to find the session fascinating.
If you're interested in learning about nanotechnology…