By @finchtalk (Todd Smith)
In 2014 and beyond Finchtalk will be contributing to Digitalbio’s blog at this site. We kick off 2014 with Finchtalk’s traditional post on the annual database issue from Nucleic Acids Research (NAR).
Biological data and databases are ever expanding. This year was no exception as the number of databases tracked by NAR grew from 1512 to 1552. In the leadoff introduction [1] the authors summarize this year’s issue and the status of the NAR index. The 21st issue includes 185 articles with 58 new databases and 123 updates. In the 1552 database repository, 193 had their URLs corrected and 24 were removed because they were deemed obsolete.
In previous posts (below) I’ve made the point that biological databases are increasing in specialization as the overall numbers increase. That trend continues. Six of the articles, highlighted in the introduction, illustrate the degree of specialization and roles different databases serve. When the databases are examined in more detail, several challenges can be noted that limit the utility of these tools. These databases include miRBASE [2], miRNEST [3], NOCODE [4], miRTarBase [5], PolymiRTS [6], and starBase [7]. All focus on non-coding RNA (ncRNA).
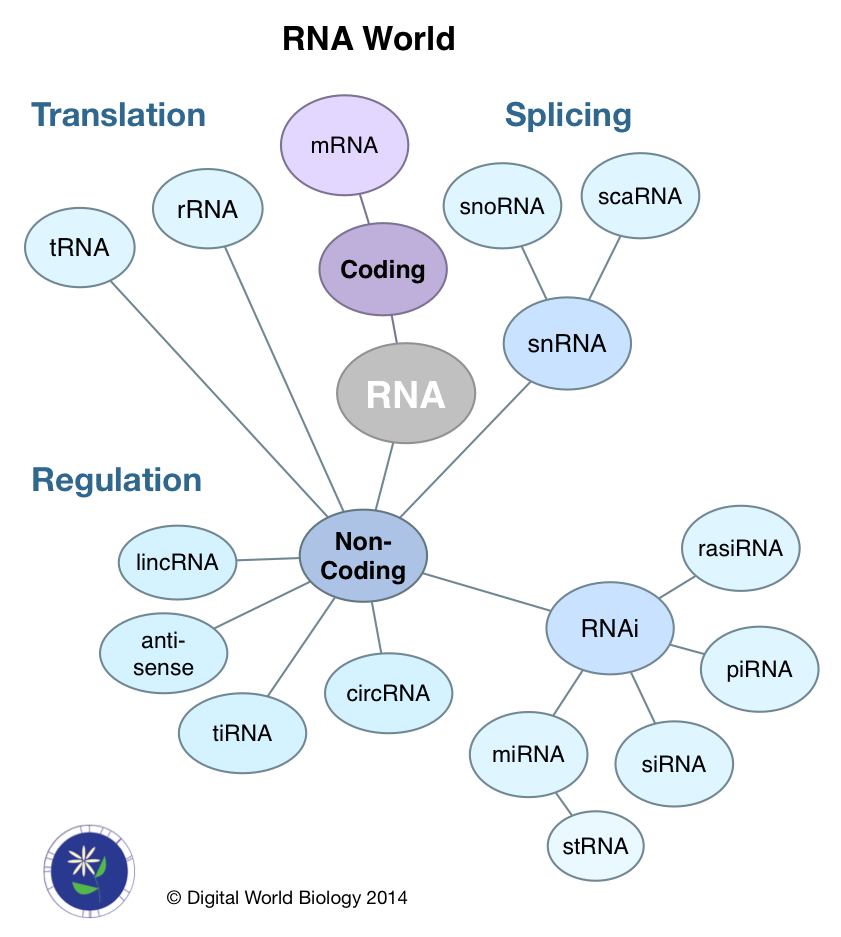 We often look at gene expression by examining the mRNA levels for genes under different conditions or in different tissues. The fact of the matter is mRNA is only a single type of RNA. The majority of RNA molecules do not code for proteins. Instead they are either involved in the process of creating mRNA (splicing), translating the mRNA into protein (rRNA, tRNA), or regulating the abundance of mRNA molecules (ncRNA). This later class is highly diverse with respect to the lengths of RNA molecules and their mechanisms of action. Four of the databases, miRBase, miRENST, miTarBase, and PolymiRTS focus specifically on micro or miRNA. NOCODE and starBase attempt to be comprehensive for all forms of ncRNA.
We often look at gene expression by examining the mRNA levels for genes under different conditions or in different tissues. The fact of the matter is mRNA is only a single type of RNA. The majority of RNA molecules do not code for proteins. Instead they are either involved in the process of creating mRNA (splicing), translating the mRNA into protein (rRNA, tRNA), or regulating the abundance of mRNA molecules (ncRNA). This later class is highly diverse with respect to the lengths of RNA molecules and their mechanisms of action. Four of the databases, miRBase, miRENST, miTarBase, and PolymiRTS focus specifically on micro or miRNA. NOCODE and starBase attempt to be comprehensive for all forms of ncRNA.
The ncRNA family is diverse and represents a next frontier in RNA research. Starting with the discovery that double stranded RNA (dsRNA) could inhibit gene expression by turning on RNA interference (RNAi) pathways [8], new RNAs were identified, micro (miRNA) and small interfering (siRNA), as essential to the RNAi pathway. Some ncRNAs are large, like lincRNAs (large intervening non-coding RNAs) [10], but most are small between 18 and 31 nt. Within in the small ncRNA group are piwi-interacting (piRNA), repeat associated small interfering (rasiRNA), small temporal (stRNA), and transcription initiation (tiRNA) RNA. Recently, new kinds of circular RNAs (circRNA) have been discovered that bind miRNAs, thus working as regulators of regulators.
How are the databases used?
 Databases help researchers study ncRNAs in two ways. First they provide ways to organize sequences and biological information (annotations) about the sequence such as the organism where the RNA was found, papers describing its discovery, algorithms used to predict its existence, and whether there are precursor forms of the active molecule. These kinds of databases can be classified as repositories and are commonly used to look up information in experiments designed to measure which RNAs are present in different tissues, at different times, or in response to environmental stimuli. For example miRBase is a collection of miRNA sequences with evidence from the literature for 206 organisms and contains 24,521 records, whereas miRNENST includes miRNA sequences that come from other databases and / or are predicted by sequence analysis programs and contains 39,122 records from 522 organisms. NOCODE is the largest repository with 148,172 records for eight organisms.
Databases help researchers study ncRNAs in two ways. First they provide ways to organize sequences and biological information (annotations) about the sequence such as the organism where the RNA was found, papers describing its discovery, algorithms used to predict its existence, and whether there are precursor forms of the active molecule. These kinds of databases can be classified as repositories and are commonly used to look up information in experiments designed to measure which RNAs are present in different tissues, at different times, or in response to environmental stimuli. For example miRBase is a collection of miRNA sequences with evidence from the literature for 206 organisms and contains 24,521 records, whereas miRNENST includes miRNA sequences that come from other databases and / or are predicted by sequence analysis programs and contains 39,122 records from 522 organisms. NOCODE is the largest repository with 148,172 records for eight organisms.
The second application is to develop an integrated resource to understand the RNA’s mechanism of action at a molecular level. Researchers create these resources to store data from experiments, but also mine the data contained to predict new kinds of interactions that can be tested in later experiments. The miTarBase, PolymiRTS, and starBase databases are examples that combine data from miRBase, other databases, literature, datasets, and predictive algorithms to create records about specific kinds of interactions and sequences at interacting sites. Each database varies with respect to the classes of RNA stored, organisms, and specific methods for identifying interactions; miTarBase (18 species, 51,460 interactions) and PolymiRTS (human and mouse, 18,514 interactions) focus on miRNA and starBase (human, mouse, C. elegans) include all ncRNA (>500,000 interactions). PolymiRTS also includes validated interactions (2944) and provides data regarding sequence variation and mutation in ncRNA function.
As a specific example, in cancer research, a scientist might want to look at miRNA expression between tumor and normal cells to see if there are miRNAs that are present in normal cells but not observed in a tumor, hence unavailable to turn off that activity of an oncogene. They would purify small RNA fragments, convert them to DNA, and sequence the products for each cell type. Next, they would compare expression profiles between the samples. Annotations from miRBase would be used assign names to the miRNAs and later look up potential functions for miRNAs that are differentially expressed between the tumor and normal cells. Using miTarBase or starBase, they could see if information about these RNAs was present and further explore how the miRNAs interact with their targets to develop hypotheses about how missing the miRNA leads to uncontrolled growth.
In some cases novel sequences can be discovered. These are either mutated forms of miRNA, or miRNAs that have not been reported in the literature. In the later case an expanded search could be conducted in miRNEST or NOCODE (to test the idea that the sequence was from a different kind of ncRNA). The researcher could also see if the sequences are present in PolymiRTS to see if their candidates have a known polymorphisms. As in the above example, the integrative resources could be used to learn about known and predicted interactions between the ncRNAs and other molecules, like the mRNAs for known cancer genes. In the case where no database matches are found, it is still possible that a completely novel, unpredicted, ncRNA could be discovered.
Clearly, these are diverse resources from a content and scientific mission perspective. They also have substantial variability regarding their technical implementations. While all strive to make their data available, only starBase provides an API for programatic access, and, at the same time is the only one lacking bulk data downloads. In the cases where bulk data is accessible, the formats vary, with tables being in tab delimited text, GFF3, BED, or other formats and sequences being commonly in a fasta format. One can also expect variable quality too. Giving the large differences between the numbers of records and how they were created, false positive data (unverifiable predictions or literature that cannot be reproduced) are expected, but levels or estimates of the number are not provided.
Organizing data in ways to create new knowledge is an important aspect of modern biology. These databases are significant in that they can provide new insights into gene regulation and biological systems. The challenge for researchers is understanding the diversity of specialized resources, application fit, and their unique methods for data delivery. These six examples focus on a common aspect of molecular biology, yet are a very diverse tip of the iceberg. The miTarBase web site alone links to an additional 23 resources that include analysis tools and other ncRNA databases, and the NAR index now references 1552 databases organized into 14 categories and 41 subcategories.
Further Reading
Previous Posts:
Small RNAs Get Smaller - http://finchtalk.blogspot.com/2009/05/small-rnas-get-smaller.html
Databases of Databases - http://finchtalk.blogspot.com/2011/01/databases-of-databases.html
Bio Databases 2012 - http://finchtalk.blogspot.com/2012/01/bio-databases-2012.html
Bio Databases 2013 - http://finchtalk.blogspot.com/2013/01/bio-databases-2013.html
Small RNAs:
ncRNA - http://nar.oxfordjournals.org/cgi/reprint/35/suppl_1/D178
snoRNA - http://en.wikipedia.org/wiki/SnoRNA
siRNA - http://en.wikipedia.org/wiki/SiRNA
miRNA - http://en.wikipedia.org/wiki/MicroRNA
piRNA - http://en.wikipedia.org/wiki/Piwi-interacting_RNA
rasiRNA - http://en.wikipedia.org/wiki/RasiRNA
stRNA - http://jcs.biologists.org/cgi/content/full/116/23/4689
tiRNA - http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3170176/
circRNA - http://en.wikipedia.org/wiki/Circular_RNA
microRNAs and Cancer - http://scienceblogs.com/pharyngula/2013/10/12/micrornas-and-cancer
References
[1] Fernández-Suárez XM, Rigden DJ, Galperin MY (2014) The 2014 Nucleic Acids Research Database Issue and an updated NAR online Molecular Biology Database Collection. Nucleic Acids Research 42(1): D1-D6.
[2] Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42(D1): D68-D73.
[3] Szcześniak MW, Makałowska I (2014) miRNEST 2.0: a database of plant and animal microRNAs. Nucleic Acids Research 42(D1): D74-D77.
[4] Xie C, Yuan J, Li H, et al. (2013) NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Research 42(D1): D98-D103.
[5] Hsu S-D, Tseng Y-T, Shrestha S, et al. (2014) miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Research 42(D1): D78-D85.
[6] Bhattacharya A, Ziebarth JD, Cui Y (2014) PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Research 42(D1): D86-D91.
[7] Li J-H, Liu S, Zhou H, et al. (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research 42(D1): D92-D97.
[9] Fire A, Xu S, Montgomery MK, et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669): 806-811.
- Log in to post comments
