digital biology
Is there a place for citizen scientists in the world of digital biology?
Many of the citizen science projects that I've been reading about have a common structure. There's a University lab at the top, outreach educators in the middle, and a group of citizens out in the field collecting data.
After the data are collected, they end up in a database somewhere and the University researchers analyze them and write papers. At least that's my impression so far.
It seems to me, that with all kinds of databases out there, on-line, there should be plenty of opportunity for both citizens and student…
"What's in a name? that which we call a rose
By any other name would smell as sweet"
- Juliet, from Romeo and Juliet by William Shakespeare
I realized from the comments on my previous post and from Mike's post that more explanations were in order.
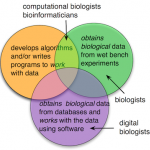
It seems we have two topics - why do we need a new name at all? and why the current names (biologist, computational biologist, bioinformatician, etc.) don't work. What really distinguishes a digital biologist from a regular, garden variety biologist? Why isn't a digital biologist a computational biologist?
So, I brought along two "show and tell…
What do you call a biologist who uses bioinformatics tools to do research, but doesn't program?
You don't know?
Neither does anyone else.
The names we use
People who practice biology are known by many names, so many, that the number of names almost reflects the diversity of biology itself.
Sometimes we describe biologists by the subject they study. Thus, we have biologists from anatomists to zoologists, and everything in between: addiction researchers, chronobiologists, epidemiologists, immunologists, microbiologists, neuroscientists, pharmacologists, physiologists, plant biologists,…
You might think the coolest thing about the Next Generation DNA Sequencing technologies is that we can use them to sequence long-dead mammoths, entire populations of microbes, or bits of bone from Neanderthals.
But you would be wrong.
Sure, those are all cool things to do, but Next Generation DNA sequencing (or NGS for short) can give us answers to questions that are far, far more interesting.
With NGS, we can look at entire transcriptomes (!!) together with the proteins that make them and the DNA modifications that help regulate them. If we compare a cell to music, a genome sequence…
What tells us that this new form of H1N1 is swine flu and not regular old human flu or avian flu?
If we had a lab, we might use antibodies, but when you're a digital biologist, you use a computer.
Activity 4. Picking influenza sequences and comparing them with phylogenetic trees
We can get the genome sequences, piece by piece, as I described in earlier, but the NCBI has other tools that are useful, too.
The Influenza Virus Resource will let us pick sequences, align them, and make trees so we can quickly compare the sequences to each other.
This is how I got the sequences that I wrote about…
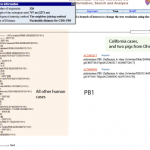
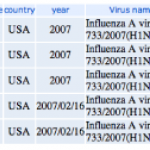
This afternoon, I was working on educational activities and suddenly realized that the H1N1 strain that caused the California outbreak might be the same strain that caused an outbreak in 2007 at an Ohio country fair.
UPDATE: I'm not so certain anymore that the strains are the same. I'm doing some work with nucleic acid sequences to look further at similarity.
Here's the data.
Once I realized that the genome sequences from the H1N1 swine flu were in the NCBI's virus genome resources database, I had to take a look.
And, like eating potato chips, making phylogenetic trees is a little bit…
I'm a big of learning from data. There are many things we can learn about swine flu and other kinds of flu by using public databases.
In digital biology activity 1, we learned about the kinds of creatures that can get flu. Personally, I'm a little skeptical about the blowfly, but...
Now, you might wonder, what kinds of flu do these different creatures get? Are they all getting H1N1, or do they get different variations? What are H and N anyway?
We can discuss all of these, but for now, lets see what kinds of flu strains infect different kinds of creatures.
Activity 2. What flu infects…
Genome sequences from California and Texas isolates of the H1N1 swine flu are already available for exploration at the NCBI. Let's do a bit of digital biology and see what we can learn.
Activity 1. What kinds of animals get the flu?
For the past few years we've been worrying about avian (bird). Now, we're hearing about swine (pig) flu.
All of this news might you wonder just who gets the flu besides pigs, birds, and humans. We can find out by looking at the data.
Over the past few years, researchers have been sequencing influenza genomes and depositing those genomes in public…
"Digital biology," as I use the phrase, refers to the idea of using digital information for doing biology. This digital information comes from multiple sources such as DNA sequences, protein sequences, DNA hybridization, molecular structures, analytical chemistry, biomarkers, images, GIS, and more. We obtain this information either from experiments or from a wide variety of databases and we work with this information using several kinds of bioinformatics tools.
The reason I'm calling this field "digital biology" and not "bioinformatics" (even though I typically use the terms as…