DNA sequencing
Scale, proportion, and quantity belong to one of the cross cutting concepts in the next generation science standards (NGSS). According to Volume 2 of the NGSS, "in engineering, no structure could be conceived much less constructed without the engineer's precise sense of scale." The authors go on to note that scale and proportion are best understood using the scientific practice of working with models.
When scientists and engineers work with these concepts at a molecular scale, new kinds of technologies can be created to advance our understanding of…
A few weeks back, we published a review about the development and role of the human reference genome. A key point of the reference genome is that it is not a single sequence. Instead it is an assembly of consensus sequences that are designed to deal with variation in the human population and uncertainty in the data. The reference is a map and like a geographical maps evolves though increased understanding over time.
From the Wiley On Line site:
Abstract
Genome maps, like geographical maps, need to be interpreted carefully. Although maps are essential to exploration and navigation they…
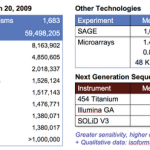
In our series on why $1000 genomes cost $2000, I raised the issue that the $1000 genome is a value based on simplistic calculations that do not account for the costs of confirming the results. Next, I discussed how errors are a natural occurrence of the many processing steps required to sequence DNA and why results need to be verified. In this and follow-on posts, I will discuss the four ways (oversampling, technical replicates, biological replicates, and cross-platform replicates) that results can be verified as recommended by Robasky et. al. [1].
The game Telephone teaches us how a…
In simple Mendelian genetics, a single change in one gene can produce a large change in mortality. The National Human Genome Research Institute (NHGRI) will be funding genomics studies on Mendelian traits using a similar strategy.
NHGRI will fund a small number of centers, dominant centers you might say, and look for large changes. The sequencing centers that will benefit are the Broad Institute, Washington University, and Baylor College of Medicine. For the next four years, the big three will be dividing $86 million a year according to a press release from NHGRI. I'm not sure what algorithms…
I had the good fortune on Thursday to hear a fascinating talk on deep transcriptome analysis by Chris Mason, Assistant Professor, at the Institute for Computational Biomedicine at
Cornell University.
Several intriguing observations were presented during the talk. I'll present the key points first and then discuss the data.
These data concern the human transcriptome, and at least some of the results are supported by follow on studies with data from the pigmy tailed macaque.
Some of the most interesting points from Mason's talk were:
A large fraction of the existing genome…
You might think the coolest thing about the Next Generation DNA Sequencing technologies is that we can use them to sequence long-dead mammoths, entire populations of microbes, or bits of bone from Neanderthals.
But you would be wrong.
Sure, those are all cool things to do, but Next Generation DNA sequencing (or NGS for short) can give us answers to questions that are far, far more interesting.
With NGS, we can look at entire transcriptomes (!!) together with the proteins that make them and the DNA modifications that help regulate them. If we compare a cell to music, a genome sequence…
One of the interesting things I learned today was that many people are calling for the genome sequences of the chimps and Macaques to be finished.
This is especially amusing because the human genome isn't quite done. We're primates, too! Why not finish our genome?
[I blame these new-found revelations on Twitter. Despite my youngest daughter's warning that only old people use Twitter, I've joined my SciBlings and taken the plunge. (you can even follow me! @digitalbio).
Now, I get to indulge my geeky tendencies while waiting in line at the grocery store. I just type #cshl and voila! I…
Genome Web's Daily Scan noted an interesting blog post today from John D. Halamka, one of the people to get his genome sequenced through the personal genome project.
I was interested to see his post since Genome Web wrote that he was discussing data standards and we have been writing quite a bit, ourselves, about data measurements for Next Gen sequencing (e.g. Next Gen-Omics) on our company blog, FinchTalk.
But Halamka didn't write about standards for data.
He wrote about standards for metadata, like family histories, and the things that are done with data after it's been collected.
All of…
A few days ago, I wrote about a cool project that some high school students did where they used DNA sequencing to identify seafood.
One question that came up from one of my commenters was how a school would start a project like this. I'm totally biased, but I think DNA sequencing (well, actually the data analysis) is one of the most interesting things that a class can do as part of a research project. These days, getting started with this kind of project, wouldn't be so hard.
Here's are some ways that I would get started:
Find an existing project where my students could collaborate and…
Two teenagers, Kate Stoeckle and Louisa Strauss, carried out their own science project over the past year. They visited 4 restaurants and 10 grocery stores and gathered 60 samples of fish and sent them off to the University of Guelph to get sequenced.
I like this story. One of my former students did a project like this for the FDA years ago, sampling fish from the Pike Place Market and identifying them with PCR. He was an intern, though. Here we have students identifying sushi on their own!
Quoting the New York Times article:
They found that one-fourth of the fish samples with…
This the third part of case study where we see what happens when high school students clone and sequence genomic plant DNA. In this last part, we use the results from an automated comparison program to determine if the students cloned any genes at all and, if so, which genes were cloned. (You can also read part I and part II.)
Did they clone or not clone? That is the question.
But first, we have to answer a different question about which parts of their reads are usable and which parts are not. (A read is the sequence of bases obtained from a chromatogram file.)
How does our data get…
This the second part of three part case study where we see what happens when high school students clone and sequence genomic plant DNA. In this part, we do a bit of forensics to see how well their sequencing worked and to see if we can anything that could help them improve their results the next time they sequence.
How well did the sequencing work?
Anyone who sequences DNA needs to be aware of two kinds of problems that afflict their results. We can divide these into two categories: technical and biological.
Technical problems are identified using quality values and the number of bases…
What happens when high school students clone and sequence genomic DNA?
Background
DNA sequencing is a wonderful tool for discovery and a great technique for getting students involved in molecular science. This fall, Bio-Rad will officially begin selling their DNA cloning and sequencing kit. Now, students across the country will have the tools in hand to begin their own projects cloning and sequencing plant genes.
Of course, without bioinformatics there's no way to know what's been cloned or sequenced.
This is where we come in. As part of an agreement with Bio-Rad, we adapted a version of…
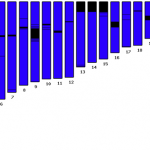
I know some of you enjoy looking at data and seeing if you can figure out what's going on.
For this Friday's puzzler, I'm going to send you to FinchTalk, our company blog, to take a look at lots of data from a resequencing experiment that was done to look for SNPs and count alleles. The graph is at the end of the post.
The graph shows data from 4608 reads (sequenced from both strands, forward and reverse). And there are some interesting patterns. Can you figure them out?
One of my colleagues has a two part series on FinchTalk (starting today) that discusses uncertainty in measurement and what that uncertainty means for the present and Next Generation DNA sequencing technologies.
I've been running into this uncertainty myself lately.
I have always known that DNA sequencing errors occur. This is why people build tools for measuring the error rate and why quality measurements are so useful for determining which data to use and which data to believe. But, some of the downstream consequences didn't really hit home for me until a recent project. This project…
Which read(s):
1. contain either a SNP (a single nucleotide polymorphism) or a position where different members of a multi-gene family have a different base?
C
2. doesn't have any DNA?
B
3. is a PCR product?
A, B, and C. All of three reads were obtained by sequencing PCR products, generated with the same set of primers.
The quality plots that I refer to are here.
Since DNA diagnostics companies seem to be sprouting like mushrooms after the rain, it seemed like a good time to talk about how DNA testing companies decipher meaning from the tests they perform.
Last week, I wrote about interpreting DNA sequence traces and the kind of work that a data analyst or bioinformatics technician does in a DNA diagnostics company. As you might imagine, looking at every single DNA sample by eye gets rather tiring. One of the things that informatics companies (like ours) do, is to try and help people analyze several samples at once so that they can scan fewer…
We have lots of DNA samples from bacteria that were isolated from dirt. Now it's time to our own metagenomics project and figure out what they are. Our class project is on a much smaller scale than the honeybee metagenomics project that I wrote about yesterday, but we're using many of the same principles.
The general process is this:
1. We sort the chromatogram data to identify good data and separate it from bad data. Informatics can help you determine if data is good, and measure how good it is, but it cannot turn bad data into good data. And, there's no point in wasting time with…
A few weeks ago, I did some "back-of-the-envelope" calculations to explain to a reader why genome sequencing costs so much.
I estimated that, if JCV's genome were sequenced at the cost advertised by university core laboratories, his genome would cost about $128 million.
That was an estimate, of course. But what did it really cost?
Genome Technology asked J. Craig himself. In the October 2007 issue of GT, JCV estimates that the cost from the first Celera human genome project (guess who?) was about $100 million and that the cost of his most recent genome project was at least $70 million.…
"Come quickly, Watson," said Sherlock Holmes, "I've been asked to review a mysterious sequence, whose importance I'm only now beginning to comprehend."
The unidentified stranger handed Holmes a piece of paper inscribed with symbols and said it was a map of unparalleled value.
Holmes gazed thoughtfully at the map, then slowly lifted his eyes and coldly surveyed his subject's beaming countenance. "You have an affinity for the ocean," said Holmes, "that you indulged to excess as a reckless youth. An experience as a medic in the military changed your life and gave you a reason to do more than…