Did you know small fragments of DNA are circulating in your blood stream?
These short pieces of DNA are left behind after cells self-destruct. This self-destruction, or apoptosis, is a normal process. In the case of fetal development, certain cells in our hands die, leaving behind individual fingers. Immune system cells leave traces of DNA behind after they’ve tackled invading microbes. DNA can also appear in the blood when people have cancer.
I had the good fortune, last Monday, to hear Matthew Snyder describe this cell-free DNA in a fascinating talk and learn why DNA in the blood can be a useful thing. It turns out that cell free DNA is a potentially useful tool for evaluating fetal health, guiding cancer treatment, and monitoring organ transplants.
According to Snyder, the use of cell free DNA for diagnosing trisomy 21, is one of the fastest growing molecular tests in the history of medicine. Some of the rapid adoption of this test is driven by pregnant women who request it.
People are interested in the prospect of using cell free DNA for other kinds of tests as well. It could be used as a biomarker to indicate the presence of cancer, or perhaps other kinds of disease.
Snyder’s research involves sequencing this cell free DNA and trying to figure out where it came from. You might think that the DNA in one person would be pretty much the same from one cell to another. And with a few exceptions, like B and T cells, that’s the case. But the DNA fragments that float around in our blood aren’t random. We can only find DNA fragments in our blood because they were hidden from hungry nucleases during the self-destruction process. Normally, those enzymes would have chopped that DNA into tiny bits.
Cell free DNA exists because the proteins that transcribe DNA and the histone proteins that package it into nucleosomes also protect DNA from being digested.
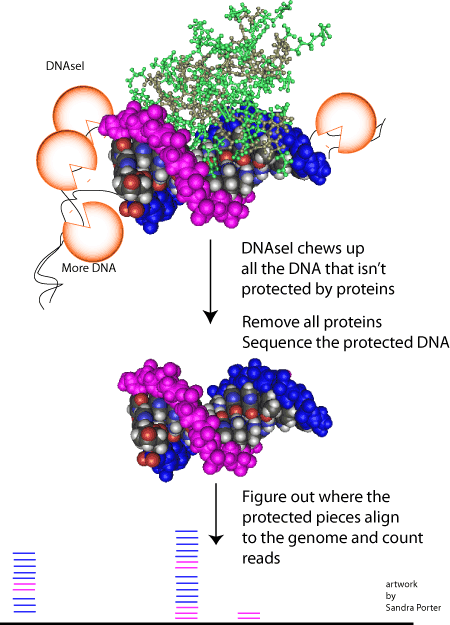 Proteins protecting DNA from digestion.
Proteins protecting DNA from digestion.
The really interesting thing, in terms of cell free DNA, is that nucleosomes and transcription factors sit on different regions of DNA in different cells. Since different bits of DNA get protected in different cells, we can sequence the cell free bits of DNA figure out where it came from. That information can tell us about a type of cancer or help us evaluate the health of multiple cell types.
 This structure has been colored by charge. The negatively charged DNA (red) is wrapped about the positively charged histone proteins. Blue represents a positive charge.
This structure has been colored by charge. The negatively charged DNA (red) is wrapped about the positively charged histone proteins. Blue represents a positive charge.
Reference:
Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016 Jan 14;164(1-2):57-68. doi: 10.1016/j.cell.2015.11.050.

This topic is so fascinating, I really enjoyed your post. The idea that this cell free DNA can act as an identifier for abnormalities that cause things like Trisomy 21 and potentially cancers could really be revolutionary. I'm surprised this topic has not gotten more attention nationally yet. After doing some research, I realized that this test is actually considered better than the old way of detecting chromosomal disorders. Check out this article if you are interested in learning more: http://well.blogs.nytimes.com/2014/02/26/new-dna-test-better-at-predict…