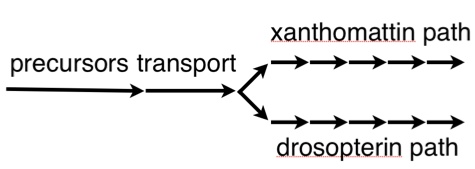
A while back, I told you all about this small piece of the biochemistry of the fly eye — the pathways that make the brown and red pigments that color the eye.
I left it with a question: if even my abbreviated summary revealed considerable complexity, how could this pathway evolve? Changing anything produces a failure or change in the result. Before I answer, let's make the problem even harder, because I love a challenge (although actually, I'm cheating — it's going to turn out that complexity is not a barrier, but an opportunity).
The pigment pathways above are far downstream: they operate in the differentiated compound eye of the fly. Long before that, there are a set of genes that have to be activated first to trigger formation of the head and eye in the larva. And this is that pathway:
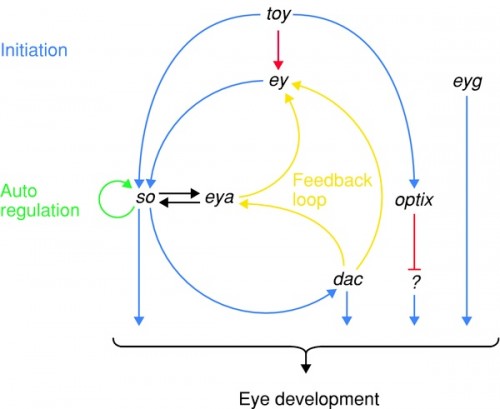
Regulatory scheme on the top of the eye developmental pathway. Twin of eyeless (toy), eyeless (ey), and possibly eyegone (eyg), three Pax genes, are master control genes on the top of the hierarchy. Sine oculis (so), eyes absent (eya), dachshund (dac), and optix are second-order transcription factors regulated by the master control genes. Note that the pathway is not linear, but rather a network interconnected by feedback loops.
At the top of the hierarchy are two genes in Drosophila, eyeless (ey) and twin of eyeless (toy). Remember, genes are named for their mutant effect, so the normal function of eyeless is to initiate eye development. These genes switch on sine oculis and eyes absent (notice the effort to find synonyms to describe genes that cause missing eyes when broken) that activate each other and feed back on eyeless to generate a robust response. Another gene, dachshund (this one named for another part of its phenotype: it makes flies with very short legs) also feeds back on eyeless.
This circuit has multiple outputs: so, dac, optix and eyg. All of these have effects further downstream, in that catch-all category labeled "eye development" here. In that broad label lie multiple processes: the pigment pathways above, but also all kinds of elaborate interactions that recruit cells to specific photoreceptor functions, that organize supporting cells, like hair cells and lenses, and that induce the neural tissue of the retina and deeper parts of the nervous system. The genes ey and toy initiate a whole deep, branching network of genes that cascade together to build the many bits and pieces of the eye.
These two master control genes, eyeless and twin of eyeless, also have a synonym. To everyone's surprise, versions of this circuit are found in all animals with eyes, and the common name for this universal regulator of eye formation is Pax6, and that's what I'll call it in the rest of this article.
And look at this! Isn't it cool? All these eyes use this same Pax6 gene regulatory network to initiate development.
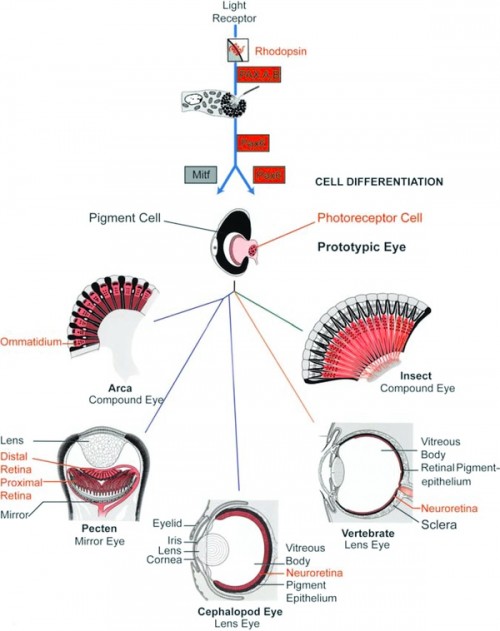
General scheme of eye evolution. The first step in eye evolution is the evolution of a light receptor molecule which in all metazoans is rhodopsin. In the most ancestral metazoa, the sponges, a single Pax gene, but no opsin gene has been found. In the cubozoan jellyfish Tripedalia, a unicellular photoreceptor has been described in the larva. The adult jellyfish has complex lens eyes that form under the control of PaxB, whereas the eyes of a hydrozoan jellyfish (Cladonema) are controlled by PaxA. We propose that from the unicellular photoreceptor cell, the prototypic eye postulated by Darwin originated by a first step of cellular differentiation into a photoreceptor cell and a pigment cell, controlled by Pax6 and MITF, respectively. From this prototype, all the more complex eye types arose monophyletically. As a mechanism, we propose intercalary evolution of progressively more genes such as lens genes into the eye developmental pathway (after Gehring and Seimiya 2010). Starting from the common prototype, the various eye types evolved by divergent, parallel, and convergent evolution, generating a magnificent biodiversity.
That's the power of a gene regulatory network. Switch on just one of the key genes, and it recruits all the downstream genes and triggers a whole series of actions to assemble a complex structure. That strange grey object to the right is a developing Drosophila wing — the dark fringe is the line of bristles that surround the leading and trailing edges of the fully-formed wing — which has had the Pax6 gene inappropriately expressed in a few cells at the base. Just switching on that one gene has led to the construction of an eye with its red pigment right there, where flies should not have eyes.
The ability to build elaborate organs with a simple switch is a reflection of the modular nature of developmental programs. It also simplifies evolution; small, simple changes can lead to dramatic novelties. Zap, one mutation can lead to an abrupt saltational change.
Now wait a moment, you will say. Suddenly plopping an eye onto a wing sounds disastrous: it really is a kind of hopeful monster, emphasis on "monster", and is almost always going to be grossly deleterious. This can't be a viable pathway for evolutionary change, can it? And you'd be right. But what about portions of a pathway? Look back up at the eye development pathway, the second figure in this article. What if you just switched on optix, one of the second-order transcription factors? Then you'd just activate some of the tools of eye construction.
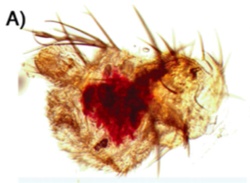
It's been done. The hideous blob to the left is the nascent antenna of a fruit fly, and optix has been inappropriately switched on…and what do you get? It activates the pigment pathway (that biochemical sequence illustrated in the first image at the top of this page), and it creates a bright red spot on the antenna. This is non-trivial; it means the precursors and transporters are all at work, and all the enzymes in the xanthomattin and drosopterin pathways are doing their job. One switch, and you get a whole hierarchy of genes producing a complex output. This could be one way new traits appear, by redeploying genes from established pathways.
Saying they could isn't the same as saying they did, of course. But here are a few examples that suggest that eye network genes have been redeployed to create morphological novelties. In Heliconius butterflies, for instance, the red spots on their wings can be traced back to embryonic patterns of expression of optix in the developing wings.
Heliconius butterflies express optix in wing epidermal cells that will produce red ommochrome pigments. A: Heliconius erato. B: Forewing and hindwing patterns from different races of H. erato (top: H. e. petiverana; bottom: H. e. erato). C: Pupal wings expressing optix mRNA in a pattern corresponding to the areas of red pigment in wings depicted in (B).
Even more dramatically, here's an extinct biting midge preserved in amber, and look at that wing: what was I saying about switching on eye genes inappropriately in the wing would be deleterious? I was wrong. This is an insect with a compound eye growing in its wing.
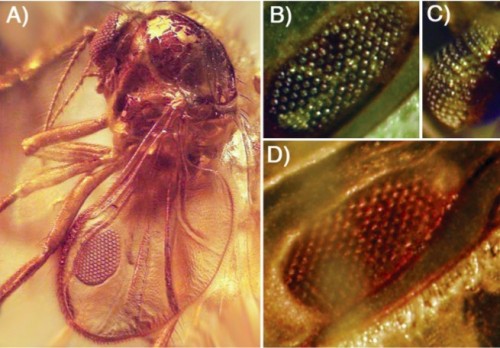
A. The extinct biting midge, Eohelea petrunkevitchi, with a unique wing organ that resembles the surface of its compound eye. B: The dorsal surface of the wing organ. C: The midge's compound eye. D: The ventral surface of the wing organ.
It's extremely unlikely that that alar eye functioned as a visual organ: any photoreceptor signals coming from a platform flapping several times a second would be hopelessly confusing. Most likely what it was was a species-specific sexual signal, like the spots on many fly wings — this one is just more elaborately structured and expensive than most. Alternatively, one hypothesis for the formation of spots on insect wings is that they are intended to resemble eyes — large eyes, far apart, making the animal look much larger to predators — so Eohelea may have just been carrying the eyespot mimicry to an extreme. Either way, building these eyes is developmentally trivial.
It may also represent a transitional state: first the initiator of a genetic cascade is co-opted and expressed at a novel time or place, and then selection can hone it down over time, adding new control points that, for instance, suppress irrelevant ommatidium formation in the alar eye while allowing the functional pigment expression to continue.
One last example: this is the Cambrian worm, Microdictyon. Notice anything unusual?
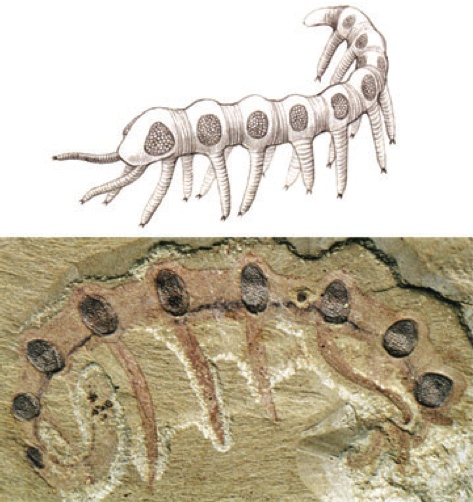
Microdictyon sinicum, a Cambrian Lobopodian fossil from Chengjiang (China) with compound eye on every annulus (segment) above every leg. (A) Reconsruction (after Bergström and Hou). (B) Lateral view.
There's a pair of eyes in the head, where you'd expect them…but all those other eyes along the sides are morphologically indistinguishable from the anteriormost pair. There is some argument about whether these structures actually are eyes, but they are definitely hexagonal arrays that closely resemble the hexagonally structured ommatidia of the compound eyes of insects. If they weren't functional eyes, it seems likely that they are at least produced by the redeployment of the structural genes of the compound eye.
And if they were functional eyes, well, that is just freakin' cool.
The bottom line, though, is that because complex developmental networks are functionally constrained — think of them as software modules that respond to molecular inputs and produce morphological outputs — their complexity is not a barrier to evolution at all, but instead provide opportunities for generating interesting evolutionary novelties.
Gehring WJ (2012) The animal body plan, the prototypic body segment, and eye evolution. Evolution & Development 14(1):34-36.
Monteiro A (2012) Gene regulatory networks reused to build novel traits. Bioessays 34:181-186.
(Also on FtB)