The field of immunology has a few quirks. I'm sure this is no different than other fields of study, but one of the most puzzling (and sometimes infuriating) of these quirks is an obsession with categorizing different types of cells. Case in point, a recent paper in Nature Immunology:
![]() A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
But before I get into that, allow me to provide some back-story.
Identifying Cells
First, I should be clear, immunology isn't alone in trying to delineate different cell types. In order to figure out how a cell or an organism works, you have to figure out how to distinguish that cell or organism from all the others. When microscopes came along, the first things people noticed were that not all cells have the same shape. For instance, microbiologists divided bacteria into bacilli, cocci and spirochetes.

(Source)
Early microscopists also pioneered the use of various stains and dyes, such as the Gram stain which is still used to classify bacteria into two major groups - Gram positive and gram negative. Early characterization of the cells in blood was no different. Red blood cells (which carry oxygen) and white blood cells (which includes everything else). Eosinophils stain positive for eosin, basophils are positive for various basic dyes. Monocytes have one nucleus, while polymorphonuclear cells seem to have many, oddly shpaed nuclei. The point of all of these characterizations is that cells with similar shape and similar properties often share the same function, and isolating the cell allows you to isolate the function.
But through the 1950's and 60's, this was still no easy feat. Many of the stains used in microscopy kill cells, which is fine if you want to look at whether or not a piece of tumor you cut out is cancerous (cancer cells often look different from normal cells), or if you want to look at the pathology of a tissue during an autopsy. But scientists remained severely hampered in their efforts to extract, isolate and identify living cells in order to study them. Then, in the late 1960's, German scientists developed a technology that would revolutionize the study of cells: Flow cytometry.
Flow Cytometry
The technology is complicated, but the principal is fairly straightforward:
1) Send a mixed group of cells through a tube one at a time.
2) Use a method of rapid analysis that allows identification of cells of interest
3) Plot positive events on a graph
or
4) Send positive cells into a different tube than negative cells
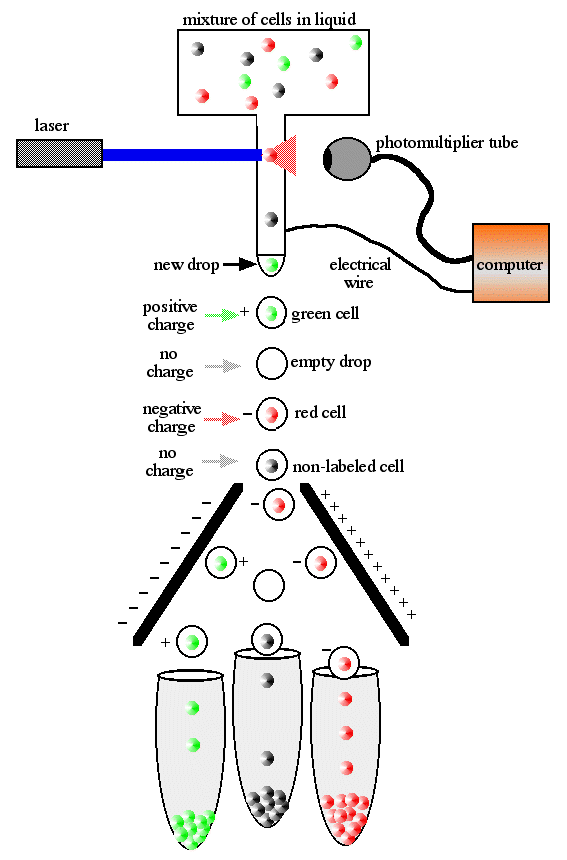
(Source)
Even using the earliest and crudest examples of this technology allowed rapid characterization of hundreds of thousands of cells at a time. Immunologists adopted this technology rapidly and effectively, due largely to two factors. First, many cells of the immune system are readily isolated from blood and are usually on their own - not bound up in large mats of other cells and connective tissue. The second reason has to do with step 2 above - immunologists had antibodies.
Antibodies
Microscopy stains served (and continue to serve) biologists well, but their application is limited. A lot of the early work on staining was haphazard and descriptive - find a new stain and see if it's useful. And as I mentioned, the staining procedure often kills cells. Antibodies, by contrast, can be raised against just about any molecule, and can be extremely specific. These days, just about every biology lab in the world makes use of antibodies as tools, but in the early days, it was the immunologists that held the key to this magic technique.
Antibodies that specifically recognized proteins on the surface of cells were churned out by the hundreds, and by attaching these antibodies to fluorescent molecules, flow cytometrists could separate cells into ever-smaller subgroups. Because of their usefulness in identifying different subgroups, these surface proteins were given systematic names, called "clusters of differentiation" or CD.

(Source)
Bone marrow and blood were no longer a mass of indistinguishable, round, mononuclear cells. Scientists could now identify stem cells from cells committed to a specific lineage. Every lineage could be sub-divided into more and more specialized groups. More importantly, these different cell-surface molecules often described something about that cell's function. CD3 was discovered early on to be a marker of T-cells, and it turns out that CD3 is an integral component of the T-cell receptor, the sine qua non of T-cells. Some T-cells express CD4, and others express CD8 - but the difference is not random. CD4+ and CD8+ cells have very different functions.
But this early and justifiable exuberance for categorization has led to some to some absurd excesses. By way of example, consider the dendritic cell. First described by Paul Langerhans in the late 19th century, these "DC's" were so named because of characteristic dendrites (from the greek for "tree", which give these cells a star-like shape. Later, immunologists learned that DC's bridge the innate and adaptive immune system by traveling from the site of infection to lymph nodes, where they present bits of infectious agents to T-cells. Early flow cytometrists also managed to find that these cells expressed high levels of the protein CD11c, and for a long time, "dendritic cell" and "CD11c positive cell" were synonymous.
Then, about 10 years ago, a new group cells were discovered. They don't look or behave anything like normal dendritic cells, they don't present antigens to T-cells, and they don't even have dendrites, but nonetheless they were called "plasmacytoid dendritic cells" (or pDC) because they express high levels of CD11c. Maybe my dismissal of this naming strikes you as pedantic, and perhaps it is, but definitions matter. People have wasted untold research dollars, experimental hours and gallons of ink trying to relate these two cell types. But I don't think they're anymore related than a DC and a macrophage.
OMG! We found a new cell!
The DC/pDC divide is one example of the sort of category-obsessed hole immunologists can fall into, but at least pDC's are a legitimately new and uncharacterized type of cell. The cell mentioned in the paper up top? Not so much. Not that you would know it from the press release.
New Cell Type Offers Immunology Hope
The discovery, published in the journal Nature Immunology, is a fundamental advance in understanding the different components of the immune system and how this system casts a net wide enough to catch all kinds of different infectious organisms.
The paper describes an NKT cell. We've known about NKT cells for over 15 years. Normal T-cells recognize fragments of proteins that are presented on the surface of cells on specialized platforms called the MHC (if you read ERV, think of the pirate-flag metaphor). NKT cells have a T-cell receptor just like their brothers, but instead of seeing bits of protein on MHC, they see bits of lipid presented on CD1. In addition, while normal T-cells have hundreds of thousands of different possible receptors, NKT cells were only known to have a very limited set of receptors. Uldrich et al merely showed that there are NKT cells that have previously unknown T-cell receptors.
This is an important step for NKT cell biology, even if only to remind the folks studying these cells that they should expand their search beyond the tiny population that they currently study. But it's hardly a paradigm shift, and this is NOT a "new" cell.
Uldrich, A., Patel, O., Cameron, G., Pellicci, D., Day, E., Sullivan, L., Kyparissoudis, K., Kjer-Nielsen, L., Vivian, J., Cao, B., Brooks, A., Williams, S., Illarionov, P., Besra, G., Turner, S., Porcelli, S., McCluskey, J., Smyth, M., Rossjohn, J., & Godfrey, D. (2011). A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties Nature Immunology, 12 (7), 616-623 DOI: 10.1038/ni.2051
So it's kind of like Heisenberg's uncertainty principle, right? Just as soon you detect/define/describe a particular sub-sub-sub type of WBC, that cell has reverted or matured to something else, by expressing or repressing a slightly different receptor or cytokine profile.
Ha! Basically. It's actually getting worse with better FACS technology - we're up to like 30 colors, and those guys have to justify their existence somehow.
Imagine if zoologists were like that; OMG, we found a new pattern within this populations of snakes, it must be a new species!
And you haven't even brought up CyTOF yet...Seriously though, even for a former NKT-maven, this was talked up a wee bit too much in the press release. I think the most interesting thing is that you can find these much more consistently in humans than the other kind, which is something towards bringing the NKTs into the fold of relevance, but still.
Hello just wanted to give you a quick heads up. The text in your post seem to be running off the screen in Ie. I'm not sure if this is a formatting issue or something to do with browser compatibility but I thought I'd post to let you know. The style and design look great though! Hope you get the issue resolved soon. Many thanks