(This is an edited excerpt from an op-ed piece I just wrote for Xconomy, posted here as I think it provides some nuance on my views on regulation of genetic testing that was lacking from my post last week.
Some context for new readers: a Congressional investigation into the direct-to-consumer (DTC) genetic testing industry last week left a sour taste in the mouths of many observers of the embryonic industry; it was a vicious, one-sided affair, starring a biased report on a "sting" operation performed by the US Government Accountability Office. Along with other recent moves by the FDA, it…
(Cross-posted to Genomes Unzipped.)
Today's US Congress Committee on Energy and Commerce hearing into the direct-to-consumer genetic testing industry was a vicious affair. Representatives from testing companies 23andMe, Navigenics and Pathway faced a barrage of questions about the accuracy and utility of their tests, made all the worse by the fact that many of the Committee's members seemed unable to distinguish between the more responsible companies in the field and the scammers and bottom-feeders. (I watched by web-cast, which I can't yet track down a copy of online; you can read the…
I mentioned last week on Genetic Future that a reporter with the Washington Post, Rob Stein, had emailed the National Society of Genetic Counsellors to search specifically for people with negative experiences of personal genomics for an upcoming article on the industry. At the time I called for personal genomics customers with positive experiences to email Stein, to ensure that he had a balanced view of the industry's impact.
Stein's article came out a couple of days ago, presumably timed for maximum impact in advance of the FDA's meeting on lab-developed tests over the last two days.…
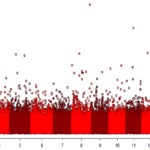
Every issue of Nature Genetics is packed full of them, and they're the basis for the risk predictions offered by every personal genomics company - but how do you make sense of a genome-wide association study? How can you tell the difference between results you can trust and those you should treat with caution?
Over at Genomes Unzipped, Jeff Barrett (who's authored more GWAS than most) explains some of the key features of these studies, and what to keep an eye out for when interpreting their results.
The first ever post on the new group blog I announced yesterday, Genomes Unzipped, is now live: it's Luke Jostins of Genetic Inference talking about the importance of sequencing for the future of personal genomics. Here's a taste:
There is a particular type of variation that genotype chips can never get at, the type of variation that most people will find most interesting: variation that is unique to you, or to your family. If you get sequenced now, about 200,000 single-base variants in your genome will never have been seen before, ever. These are likely to include changes that modify…
I'm pleased to announce the beta launch of a new group blog on personal genomics, Genomes Unzipped.
I've been working with a group of scientific colleagues and fellow bloggers on this project for quite a while now. Some of the group members will be familiar to regular readers: Dan Vorhaus from Genomics Law Report, Luke Jostins from Genetic Inference, and Caroline Wright from the PHG Foundation. Others are new to blogging, but have backgrounds in genomic analysis, statistical genetics and other fields that allow them to bring valuable insight into the scientific, ethical and social…
I've just been forwarded an email that was originally sent to the National Society of Genetic Counsellors email list on behalf of a reporter with the Washington Post:
Dear NSGC Members,
Washington Post reporter Rob Stein has interviewed NSGC President Liz Kearney on the subject of regulation of genetic testing. Rob would also like to interview patients who have experienced DTC testing. Rob is particularly interested in speaking with patients in the D.C. area who have had a bad experience with DTC testing or who sought the help of a genetic counselor to order the correct test and/or…
As an addendum to my previous post on the controversial "longevity genes" study, you should go and check this out. It's a post on the blog of personal genomics company 23andMe, and it's a pretty impressive piece of scientific dissection of the longevity GWAS paper - in addition to detailing a variety of methodological problems with the study, the authors actually used the 23andMe database to look at the predictive value of the longevity GWAS algorithm on their own customers:
We took a preliminary look in our customer data to see if the proposed SNP-based model described in…
When an article was published in Science last week reporting that DNA samples from exceptionally long-lived individuals differed detectably from those of normal individuals, it got plenty of positive attention from the mainstream media. However, the buzz from experts was rapid and telling: my colleagues in the statistical genetics community weren't excited about the results, but immediately, profoundly skeptical.
For people who've spent years doing genome-wide association studies (GWAS), several things stand out as unusual from this paper: the very large effect sizes of the identified…
An excerpt from an article I co-wrote for Xconomy with Genomics Law Report's Dan Vorhaus - link to the full article below.
Are you ready for consumer genetics? Is your government?
Recent announcements of federal investigations into the budding direct-to-consumer (DTC) genetic testing industry suggest that authorities are preparing to increase regulation of companies offering consumers access to their own genetic data. However, rather than rushing in to clamp down on the industry, regulators should slow down and focus, first, on understanding this complex field.
An increasing number of…
In October last year I reported on a presentation by direct-to-consumer genetic testing company 23andMe at the American Society of Human Genetics meeting in Honolulu, in which the company described results of genetic association studies performed using combined genetic and survey data from their customers. The results of their study include replication of several known associations for traits like hair colour, eye colour and freckling, as well as the discovery of previously unpublished associations for things like asparagus anosmia (the ability to smell urinary breakdown products after eating…
Blogging time has been pretty scarce for me lately, mainly due to the impending submission of the 1000 Genomes Project pilot paper (more on my involvement in that project later). Sadly, personal genomics has not done me the favour of sitting still while I'm busy. Here are some of the more interesting recent bits and pieces from the personal genomosphere:
Speaking of the 1000 Genomes Project, the consortium has formally announced the release of data from its three pilot projects. The pilot data include low-coverage whole-genome sequences from 180 individuals, high-quality whole-genome…
It looks as though the FDA is swooping down on the direct-to-consumer genetic testing industry in a serious way, sending formal letters to five companies informing them that their tests will be regulated as medical devices:
WASHINGTON -- The Food and Drug Administration is issuing regulatory letters to five genetic test makers, the first sign that the government is cracking down on companies that claim to use DNA samples to predict inheritable diseases.
The FDA letters notify each company that their tests are considered medical devices and therefore must be federally approved as safe and…
I posted yesterday on a serious incident at 23andMe's sample processing lab, LabCorp, that resulted in the wrong data being sent to up to 96 customers. The company has just posted a blog entry explaining the cause of the problem and the approaches being taken to ensure it doesn't happen again.
As several commenters had speculated, the cause of the problem appears to have been the flipping of a 96-well plate (a solid block of plastic containing 96 separate samples in individual wells) by 180 degrees, resulting in all of the samples being in the incorrect locations for downstream analysis.…
Some worthwhile recent links from the world of personal genomics:
A great piece in Newsweek by Mary Carmichael summarising the recent regulatory furore over direct-to-consumer genetic testing, and the potential implications for the industry.
Emily Singer has two articles at MIT Technology Review summarising important messages from the Consumer Genetics Conference last week. Firstly, early results from the Coriell Personalized Medicine Collaborative suggest that risk information from individual genetic test results is a stronger motivator for healthy behaviour than risk prediction from…
Personal genomics company 23andMe has revealed that a lab mix-up resulted in as many as 96 customers receiving the wrong data. If you have a 23andMe account you can see the formal announcement of the problem here, and I've pasted the full text at the end of this post.
It appears that a single 96-well plate of customer DNA was affected by the mix-up. This resulted in incorrect results being sent to customers, with alarming consequences in some cases; one mother posted on the 23andMe community about her distress upon discovering that her son's results were incompatible with the rest of the…
Jay Flatley, CEO of sequencing giant Illumina, announced at the Consumer Genetics Conference today that the company had reduced the price of its retail whole-genome sequencing service. At $19,500 this still isn't in the realm of an impulse buy for most of us, but it's a long way down from the $48,000 that Illumina offered at the launch of its service, and more than an order of magnitude below the $350,000 price paid for the first ever retail genome.
Illumina's press release notes that bulk orders of 5 or more genomes drop the price to $14,500 per genome, and "[i]ndividuals with…
The brief Golden Age of direct-to-consumer genetic testing - in which people could freely gain access to their own genetic information without a doctor's permission - may be about to draw to a close. In a dramatic week, announcements of investigations into direct-to-consumer genetic testing companies by both the FDA and the US Congress have sent the personal genomics industry into a spin, and it is still impossible to say exactly which way it will be pointing once the confusion passes.
I've been frustratingly unable to find the time to cover the developments as they happened due to other…
I've been quiet for the last two weeks, largely due to some feverish last-minute analysis in the lead-up to this year's Biology of Genomes meeting at Cold Spring Harbor Laboratory, where I spoke in (and co-chaired) the Genetics of Complex Traits session.
Long-term readers may recall that I sparked off a minor controversy at last year's meeting by writing several blog posts summarising presented work. While I deliberately steered clear of discussing unpublished or contentious work, basically focusing on the "big picture" messages emerging from the sessions rather than the technical details, I…
A paper just released in the Lancet describes a thorough and integrated approach to squeezing as much clinically relevant information as possible out of a genome sequence. However, despite a state-of-the-art clinical interpretation pipeline, the major message from the paper is just how far we still have to go before we can make full use of our genetic information.
The paper is based on the genome of Stephen Quake (right), which was sequenced using the single-molecule platform developed by Helicos (I wrote about Quake's genome publication at the time). This is a rather curious choice: of all…