When I started blogging, I never (EVAH!) thought I would describe the biology of E. coli with a Tolkein poem:
All that is gold does not glitter,
Not all those who wander are lost;
The old that is strong does not wither,
Deep roots are not reached by the frost.
From the ashes a fire shall be woken,
A light from the shadows shall spring;
Renewed shall be blade that was broken,
The crownless again shall be king.
I'm referring, of course, the spinach outbreak strain of E. coli O157:H7. You know, this one:
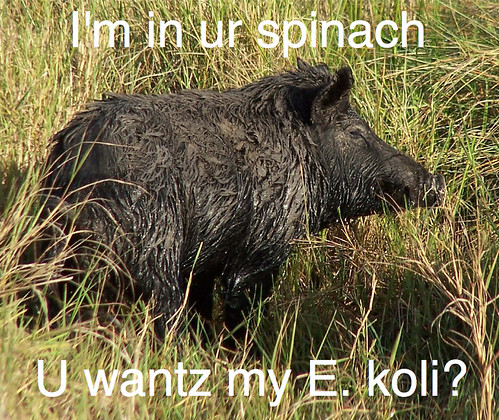
It turns out that this particular strain of E. coli O157:H7 is a member of a clade (a set of strains descended from the same common ancestor) that causes severe disease far more often than other E. coli O157:H7 strains, known as clade 8 (although it would be either far cooler or far nerdier to refer to this as the 'Aragorn' clade).
What's interesting about clade 8 is that it's the oldest clade of E. coli O157:H7, even though it didn't start causing outbreaks until 2006. In other words, the oldest group of E. coli O157:H7 has reemerged as a really nasty pathogen. Here's one way of looking at it:
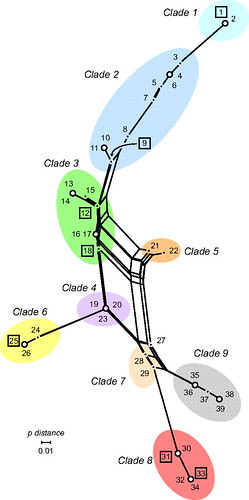
(from here)
Clade 9, in gray, is the oldest clade, and clade 8 is its closest relative. Here's another way of depicting this:
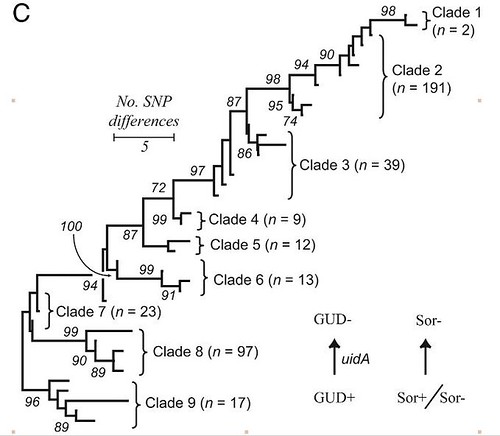
(from here)
So what makes clade 8 so special? Many clade 8 strains have a phage (bacterial virus that results in the production of a toxin known as stx2c. While other clades of E. coli O157:H7 contain the occasional stx2c bearing strain, almost sixty percent of clade 8 E. coli O157:H7 have this gene. However, while this gene might explain the severity of disease, it doesn't explain why clade 8 is doing so well, since it's primary ecological niche isn't making human sick, but living in animals. Within more resolution (i.e., more genomic data for each strain), we should be able to estimate when stx2c was acquired, and if this event (or events) are associated with other genetic changes.
Finally, because I can:


I don't get why the ascending numbers are older clades. When you discover a new one, will you give it a zero or negative number? Or will you shift them all up? If you do that then having to find every instance you ever used the old numbers and correct it. An addition would make the old list completely useless as opposed to slightly useless. As a layman I'm just confused about the logic of the naming and ordering system here...
So let me see if I've got this straight: Clade 8 is a hobbit and Clade 9 is a Klingon, and E. coli is the Millennium Falcon?
spayced,
I have no idea why the oldest clade was assigned the number nine, and not one.
"All that *glisters* is not gold."
It's Tolkien, not Tolkein!
There is no such thing as "the oldest" strain of any living organism. We are all equally old. As you note, the shiga toxin genes (such as stx2c) are carried by a phage. But as far as I can tell, we don't know yet how often this phage jumps from one strain of E. coli to another. The phage integrates into the bacterial chromosome, similarly to the way a retrovirus integrates into a eukaryotic host chromosome. How often the phage is found in the exact same chromosomal location within a particular strain of O157:H7 E. coli will tell us something about how often it moves in and out of the strain.
The I have no idea why the oldest clade was assigned the number nine, and not one ends
thanks