An article in Emerging Infectious Diseases describes a joint collaboration between the CDC and Mexican health authorities that built a system to monitor the spread of Salmonella through the food chain and into people. One finding shocked me.
The authors examined four Mexican states, and the carriage of Salmonella in asymptomatic children ranged from 1.9% to over 11 percent. That's a lot of kids with Salmonella who are not exhibiting any symptoms:
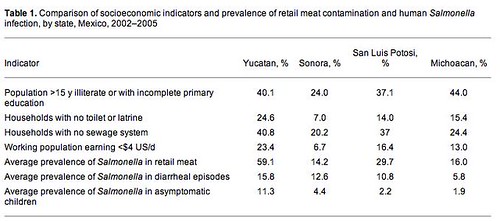
Salmonella carriage was strongly correlated with contamination of meat by Salmonella* and by overall poverty. While the healthy kids had fewer of the 'nasty' Salmonella types (Enteriditis and Typhimurium) as compared to food animals, they were still found in asymptomatic children. It's not clear why this is the case. Several explanations present themselves:
- These Salmonella were associated with a previous bout of disease that was not recorded ('healthy' was defined as having no diarrhea in the previous week), and these strains persist at a density below the threshold for causing disease.
- Mexican children acquire immunity to Salmonella, allowing for stable coexistence.
- These asymptomatic strains are less able to cause disease, and are essentially commensals.
Now, if there were only a biologist in a position to sequence some of these genomes to get at these questions....
*The table is a summary. Within the poorest communities, food contamination was associated with asymptomatic carriage.
Cited article: Zaidi MB, Calva JJ, Estrada-Garcia MT, Leon V, Vazquez G, Figueroa G, et al. Integrated food chain surveillance system for Salmonella spp. in Mexico. Emerg Infect Dis [serial on the Internet]. 2008 Mar [date cited]. Available from http://www.cdc.gov/EID/content/14/3/429.htm

Sequencing seems overkill to me - microarray analysis would do the trick - probably even just looking at Salmonella PaI presence or absence by Southern blot or PCR would start down the road. I can't believe there isn't a epidemiologist doing something like this right now...or did I miss a joke here?
Microarray/Southern/PCR only works if these Salmonella are commensal because they lack something pathogenic Salmonella have. But if they are commensals because of SNPs or because of the gain of genes, then you're going to need to sequence the genome.
(Yes, yes, you could use a tiling array for SNPs, but at that point you might as well sequence the whole genome to pick up genes that aren't on the array.)
That´s something which i always wanted to know!
Asymptomatic fecal shedding of Salmonella after a bout of clinical disease is VERY common and in humans (and livestock) lasts for a mean time of 5 weeks post infection. In livestock this can be much, much longer and can be intermittent depending on numerous factors. Are they commensals in people then.. ?
There are, in fact, species in which the bad Salmonella you refer to are commensals already- primarily chickens, where Typhimurium and Enteritidis generally do not cause disease.
Microarray analysis for genomic content probably isn't the best way to determine the genetic differences between commensal typhimurium and pathogenic Typhimurium (if there are any differences)- genome sequence or tiling array would be better. This is because it is known from sequenced Typhimurium isolates (of which there are 3 or 4), that there isn't much difference between them in terms of the presence or absence of genes- and you can't pick up SNPs (which may be more important in this case) by conventional ORF array....
Pathogenicity in Salmonellae is well known to associate with the presence of (I think 2) pathogenicity islands (there might be more). So simple genomic analysis ought to be a good starting point (something like PFGE patterns based on the known S. typhimurium/enteriditis genome sequences. You wouldn't need to worry about things like SNPs, I don't think - based on lots of other studies in lots of other bugs, there's probably a single clonal , source for the commensal population, assuming it exists. Of course, it could be a host factor or even a gut microbial population issue (i.e. some other gut bug is keeping them from causing diarrhea).
Yes, there are more than two- but we will just deal with the two major ones.
It has been shown that all Salmonella possess SPI-1, one of the major pathogenicity islands.(representatives of each species, and subspecies were tested, as well as members of the Salmonella reference collections). This pathogenicity island is thought to have been acquired at the split between Salmonellae and E. coli (100- 150 M years ago). The second pathogenicity island (SPI-2) is reportedly present in all Salmonella enterica species Enterica (this contains basically every serotype you would ever care about - in fact all of the serotypes we would care about pretty much are in a subdivision of this species called subspecies I Enterica)- and NOT present in the other species of Salmonella- S. bongori.
The nomenclature is unbelievably confusing. But the bottom line is you can't use the presence or absence of spi-1 and spi-2 to determine anything- because they are supposedly present in pretty much everything (with the exception of bongori) that is in the genus Salmonella.
This is a simple appearing problem which is actually very very complex!
I think the first step in figuring it out is to use a broad genotyping strategy to see if there is a clonal isolate that has emerged (can happen lots of ways, right?) I'm not a salmonellologist, but it seems to me that the definition of a PI is that it is there in pathogenic strains, but not in others (as an aside, I know that the nomenclature is a mess, here and in other pathogen systems!). However, my feeling is that either a clone has arisen that competes well in the gut without causing disease (displacing pathogenic salmonellae) or something else entirely is going on not directly related to the salmonella itself). My (probably somewhat inflated) $0.02.
What I was trying to say- and perhaps I am not expressing myself very well- is that possession of these two pathogenicity islands is what makes Salmonella enterica (which includes almost ALL salmonellae), Salmonella enterica- as far as we currently know.
The very same Typhimurium isolate that is a pathogen of people and calves and mice, is simultaneously a commensal of chickens. There was no loss/gain of genes (or pathogenicity islands) or generation of SNPs in Typhimurium in order for this to happen.
My guess with the Mexican children is that because there is a high prevalence of salmonellosis- and many of these children are shedding for prolonged periods after a period of clinical disease. As I said in an earlier comment- fecal shedding of Salmonella after a period of active disease can last for a mean time of 5 weeks in people- and if you look at this study I believe the judged children asymptomatic shedders as long as they hadn't had diarrhea in the preceding 7 days (or something like that).
I didn't realize that the SPIs were in all enterica isolates - kind of makes them not PIs, don't you think :) Anyway, I tend to agree with you, although the level of asymptomatic shedding is a lot higher in the one group (stats? the table didn't say) leading to the possibility of a population level difference (either in bacteria or people). I hadn't thought of your suggestion - that they had a higher rather than lower incidence of disease, that results in higher numbers of asymptomatic shedding children after infection. Of course, I am way, way out of my area of specialty, so I'm really just guessing. It sounds like an interesting epidemiological area of inquiry, though!