next-generation sequencing
In the last century infant mortality has declined precipitously in the Western world, thanks in large part to the development of antibiotics and vaccination. Yet as the suffering and death from infectious disease has reduced, the burden from genetic disease has become proportionately greater: currently around 20% of all infant deaths in developed countries are a result of inherited Mendelian (single-gene) disorders.
What can be done to reduce this burden? Increasingly sophisticated methods for detecting disease in embryos during pregnancy will help, and these have recently taken another step…
Software company 5AM Solutions has just launched a neat little FireFox plug-in for customers of consumer genomics company 23andMe.
The idea is very simple:
Download your raw data from 23andMe (or use one of the files from me or my colleagues at Genomes Unzipped);
Install the plug-in from here and point it to your 23andMe data;
Browse to a website discussing one of the genetic variants included on the 23andMe chip, and you'll see highlights around the rsID of any variant on the page (rsIDs are unique codes assigned by dbSNP to most of the common variants targeted by…
As part of his Gene Week celebration over at Forbes, Matthew Herper has a provocative post titled "Why you can't have your $1000 genome". In this post I'll explain why, while Herper's pessimism is absolutely justified for genomes produced in a medical setting, I'm confident that I'll be obtaining my own near-$1000 genome in the not-too-distant future.
Matt's underlying argument is that while sequencing costs will continue to drop, obtaining a complete genome sequence that is sufficiently accurate for medical interpretation will require additional expenses (increased sequence coverage to…
As part of his Gene Week celebration over at Forbes, Matthew Herper has a provocative post titled "Why you can't have your $1000 genome". In this post I'll explain why, while Herper's pessimism is absolutely justified for genomes produced in a medical setting, I'm confident that I'll be obtaining my own near-$1000 genome in the not-too-distant future.
Matt's underlying argument is that while sequencing costs will continue to drop, obtaining a complete genome sequence that is sufficiently accurate for medical interpretation will require additional expenses (increased sequence coverage to…
Late last week I stumbled across a press release with an attention-grabbing headline ("The Causes of Common Diseases are Not Genetic Concludes a New Analysis") linking to a lengthy blog post at the Bioscience Resource Project, a website devoted to food and agriculture. The post, written by two plant geneticists, plays a tune that will be familiar to anyone who has encountered the rhetoric of GeneWatch UK: basically, modern genomics is pure hype perpetuated by scientists seeking grant money and corporations seeking to absolve themselves of responsibility for environmental disasters. …
A reminder to anyone who reads my other blog Genomes Unzipped that we have a reader survey underway there now, which includes some questions about genetic testing experiences and attitudes towards genetics. We're closing the survey to responses this weekend, so if you're an Unzipped reader but haven't had a chance to fill in the survey, please do so now.
Update 30/11/10: 23andMe has extended their 80% discount until Christmas, without a need for a discount code.
Personal genomics company 23andMe has made some fairly major announcements this week: a brand new chip, a new product strategy (including a monthly subscription fee), and yet another discount push. What do these changes mean for existing and new customers?
The new chip
23andMe's new v3 chip is a substantial improvement over the v2 chip that most current customers were run on (the v2 was introduced back in September 2008). Firstly, the v3 chip includes nearly double the…
Back in June I launched a new blog, Genomes Unzipped, together with a group of colleagues and friends with expertise in various areas of genetics. At the time I made a rather cryptic comment about "planning much bigger things for the site over the next few months".
Today I announced what I meant by that: from today, all of the 12 members of Genomes Unzipped - including my wife and I - will be releasing their own results from a variety of genetic tests, online, for anyone to access. Initially those results consist of data from one company (23andMe) for all 12 members; deCODEme for one…
More huge news in the sequencing industry, following on from the public share offer from Pacific Biosciences - relative newcomer to the field, Ion Torrent, has just been bought by Life Technologies for an impressive US$375 million in cash and stock, with an option to increase by a further US$350 million if "certain technical and time-based milestones" are met by the end of 2012.
Ion Torrent made a splash with its launch at this year's Advances in Genome Biology and Technology meeting in February (here's my coverage from the meeting). The company has developed a sequencing technology…
The long-awaited public stock offer from third-generation sequencing technology company Pacific Biosciences has finally arrived (here's the SEC filing, and coverage from Matthew Herper and GenomeWeb).
PacBio has already raised almost US$400 million in venture capital, and aims to increase this by up to US$200 million from its share offering. The sheer scale of these figures gives you a sense of just how much money is being thrown around in the race towards ever cheaper, faster and more accurate DNA sequencing machines. Of course, whether investors will decide to throw more money in PacBio's…
The first ever post on the new group blog I announced yesterday, Genomes Unzipped, is now live: it's Luke Jostins of Genetic Inference talking about the importance of sequencing for the future of personal genomics. Here's a taste:
There is a particular type of variation that genotype chips can never get at, the type of variation that most people will find most interesting: variation that is unique to you, or to your family. If you get sequenced now, about 200,000 single-base variants in your genome will never have been seen before, ever. These are likely to include changes that modify…
Jay Flatley, CEO of sequencing giant Illumina, announced at the Consumer Genetics Conference today that the company had reduced the price of its retail whole-genome sequencing service. At $19,500 this still isn't in the realm of an impulse buy for most of us, but it's a long way down from the $48,000 that Illumina offered at the launch of its service, and more than an order of magnitude below the $350,000 price paid for the first ever retail genome.
Illumina's press release notes that bulk orders of 5 or more genomes drop the price to $14,500 per genome, and "[i]ndividuals with…
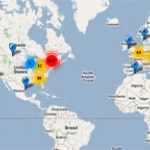
Nick Loman (of the University of Birmingham, and the Pathogens: Genes and Genomes blog) has a post updating us on his survey of second-generation sequencing machines around the world. Loman's results are also available in the format of a handy Google map (see left).
The take-home messages based on 669 machines in the database: Illumina continues to utterly dominate the second-gen market, with competing short-read platform SOLiD squabbling for scraps with Roche's 454. That's a pretty poor outcome for SOLiD, which has failed to gain traction in the market despite having the full force of…
Zoe McDougall from Oxford Nanopore points me to a press release from Illumina announcing a new era of celebrity genomics:
Illumina, Inc. (NASDAQ:ILMN) today announced that it has sequenced the DNA of American actress Glenn Close, the first publicly named female to have her DNA sequenced to full coverage. The service was completed in Illumina's CLIA certified and CAP accredited laboratory utilizing Illumina's Genome Analyzer technology and following the established process shown at http://www.everygenome.com/. Ms. Close's DNA was sequenced to an average depth greater than 30 fold, providing…
Dan Koboldt has a very nice recap of the various sequencing technologies presented at last week's Advances in Genome Biology and Technology meeting. I totally agree with his central point:
Something had been bothering me about the sequencing-company presentations this year, and I finally realized what it was. During AGBT 2009, every player was gunning to take over the world. This year it seems like every sequencing platform has a niche in mind.
The recent proliferation of sequencing technologies - each with their own characteristic profile of strengths and weaknesses - has been bewildering…
The main theme of this year's Advances in Genome Biology and Technology meeting should come as no surprise to regular readers: sequencing. Generating as many bases of DNA sequence as quickly, cheaply and accurately as possible is the goal of the moment, and the number of companies jostling to achieve that goal is growing rapidly.
The meeting saw impressive performances from established players in the field, especially Illumina: their new HiSeq 2000 instrument seems to have dug in as the platform of choice for generating vast amounts of high-quality short-read data. Life Technologies…
Stephen Turner from Pacific Biosciences gave a dramatic presentation this afternoon launching PacBio's new third-generation sequencing instrument. The room was packed for the seminar, with a palpable buzz, and Turner's presentation was preceded by a theatrical introduction from PacBio CEO Hugh Martin.
The crescendo of Turner's presentation was the unveiling of a video showing the new (and enormous) PacBio instrument, which has been tucked away in a room here at AGBT and revealed to a trickle of VIPs (including Bio-IT World's Kevin Davies) - if that's the kind of thing you're into,…
I've been remiss in blogging from the Advances in Genome Biology and Technology meeting here in Marco Island, Florida, primarily due to some panic-stricken last-minute changes to the slides for my own presentation last night.
Fortunately the conference has been extremely well-covered by others: Sanger colleague Luke Jostins has blog posts up summarising day 1 and day 2 of the meeting; Dan Koboldt from MassGenomics has his first impressions and a review of the cancer genomics session; and Anthony Fejes is continuing the tradition of publishing extensive notes on every talk he attends. …
I'll be at the Advances in Genome Biology and Technology meeting in Marco Island, Florida for the next week, soaking up sun and genomics, keeping my eye out for the anticipated major announcements from sequencing companies and researchers, and quietly panicking about my presentation on Thursday. You'll hear more about the meeting from me and the other bloggers there - Luke Jostins, David Dooling, Dan Koboldt and Anthony Fejes - over the next week.
It's amazing to think that it was at the same meeting in 2009 - just one year ago - that Complete Genomics emerged dramatically from stealth mode…
The big news from the JP Morgan investment conference today is the announcement of a brand new shiny sequencing machine from Illumina, the HiSeq 2000. The new machine boasts an impressive set of statistics, and looks likely to gradually replace Illumina's GAIIx as the workhorse of most modern sequencing facilities.
So, how excited should we be?
Let's be clear about this up front: this new machine, while impressive, represents an incremental advance rather than a dramatic technological leap forward. This is still second-generation sequencing, generating relatively small snippets of DNA…