Last Sunday I was on a panel at the American Society for Microbiology meetings. I'll post a slidecast of the talk in a few days, and get into the topic of the panel in more detail then.
Since I was presenting, I got a free pass to the rest of the conference, which was fascinating. I'm a macrobiologist by training and by avocation; I worked one summer in a fruit fly lab studying Wolbachia infections, and quickly moved on to animals one can see and touch. So I was curious to find out what microbiologists spend their time talking about.
What struck me most was how few of the techniques people were using would have existed, or at least been widely available, when I was in grad school. Many of the talks I attended involved techniques in metagenomics, a subject I had to do some quick research on from the audience just to follow along.
Metagenomics takes the sort of DNA sequencing that crime labs use for DNA fingerprinting, and it amps it up. Rather than trying to identify a single individual or species from its unique DNA, metagenomic techniques let researchers grab a sample with hundreds or thousands of microbial species. They'll take a drop of water, or a swab from the side of a hoatzin's stomach, make lots of copies a pre-determined stretch of DNA present in all of the species present, sequence all of those stretches, and then compare them to know species to assess the diversity within that droplet.
The amount of data which comes from a droplet can be staggering. Most remarkably, most of the studies I saw involved doing that analysis hundreds of times. In one case, researchers took swabs of bacteria from dozens of babies mouths, from behind their ears, and from their poop, to help them track how the microbial world inside and around babies change after birth, and to trace the effects of antibiotic treatments on that world. To do that, they had to take three such sets of samples from every baby, once a month, for years. Each month, each baby gave them 9 samples, the mother contributed several more, and the team conducted a metagenomic survey of bacterial diversity on each of them. Each of those nine analyses per baby, per month, probably involved more genomic data than was represented by all the papers in a given volume of The Journal of Mammalogy from ten years ago. And these researchers did that for dozens of babies, every month, for years.
In another case, researchers curious about the unusual digestive system of the hoatzin performed metagenomic analyses to find out how bacteria work in the unique foregut that lets the bird break down leaves. So they took samples of bacteria from the wall of the gut, others from the midst of the mass of digesting leaves, and others from further back in the gut. Another researcher used metagenomics to identify new kinds of viruses affecting the microbes of Yellowstone's hotsprings. Another used these tools to track antibiotic resistance in pets. Thanks to these tools, the researchers can examine bacterial ecosystems on a scale unimaginable only a few years ago. They can see how the bacteria on your hands and those in your gut and your nose compare, and see how those bacterial ecosystems compare to those in uncontacted populations in the Amazon, and how those bacteria are passed on to your children, or to and from your pets. They can understand how different species of microbes relate to one another in ways that were never possible before, changing how scientists think about this invisible world.
It's unfathomable how each of these authors could stand on stage and not be struck dumb by how quickly their field has transformed.
And it didn't stop there. Metagenomics involves looking at a short chunk of DNA, one variable enough to mark unique bacterial lineages, but not so large as to be impractical to sequence. But in many of the talks, researchers indicated that the followup research would involve sequencing genomes of some of the interesting bugs they'd found. Sometimes they were doing this to identify previously unknown species, but other times they just wanted to identify antibiotic resistance and see how the antibiotic resistance of several strains of a single bacterial species compared.
Again, these researchers tossed off the idea of running whole-genome sequences – sequences, plural – as if it was a chore to be assigned to a particularly dull new grad student. The first bacterial genome was sequenced in 1995, and it was published in Science and given the honor of the cover illustration. Less than 20 years later, it's routine.
The reasons for this transformation are fairly straightforward. Over the last 5 years, the cost of sequencing a genome has dropped precipitously, far faster than computer speeds increase (Moore's law says computer speeds double every 18 months):
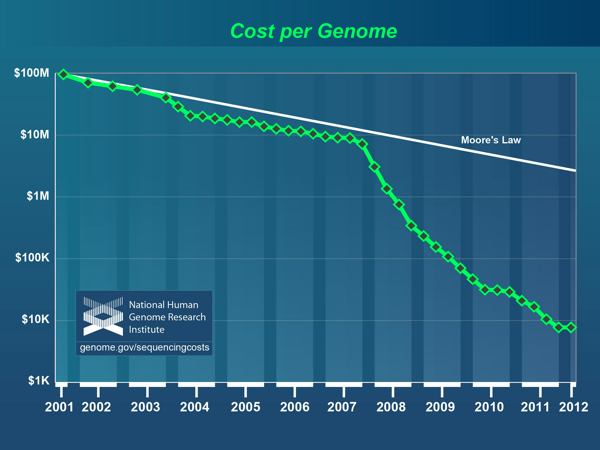
That graph, from the National Human Genome Research Institute, shows how what was once a massive undertaking, an effort that involved the combined efforts of thousands of people around the world, new inventions and years of sustained effort, now costs less than $10,000, and can be completed in a matter of hours.
Before I went to the conference, I joked with people that I'd be sure to bring my microscope with me. Boy would that have marked me as an old fogey.

Have you been in a Micro class lately? Last year, due to the out break of E. coli in Germany, they were able to sequence the entire genome in a matter of days! its crazy quick now! Metagenomics is also a common topic for all my Micro classes too! funny, I find it hard to believe these classes could exist with out advanced molecular genetic techniques!