There aren't many reports of 14 year-olds making scientific contributions. Even in the field of astronomy, Caroline Moore, the youngest person to discover a supernova, is a bit unusual.
This supernova comes from Astronomy Picture of the day. Photo credits: High-Z Supernova Search Team, HST, NASA
HT: National Science Foundation
The National Science Foundation reported that Moore has found an supernova candidate that may be as unusual as she is.
"It's really a strange supernova," said Moore. "A supernova is a huge explosion deep in the core of a star, whereas a nova is an explosion on the…
One of my all time favorite books is South: The Last Antarctic Expedition of Shackleton and the Endurance (The Explorers Club Classic) . It's an amazing adventure and an incredible story. It's a comfort to know that any challenges I face will be easier than those conquered by Shackleton.
Luckily, traveling to Antarctica these days is far less hazardous. And we're glad of that because we'd really to send one of our favorite bloggers there as our surrogate eyes and ears.
We're also glad because the we won't have to worry about her getting eaten by polar bears.
Help send Grrl to the far south,…
For those of you who may have been wondering where I've been, these past few weeks have seen me grading final projects, writing a chapter on analyzing Next Gen DNA sequencing data for the Current Protocols series, and flying back and forth between Seattle and various meetings elsewhere in the U.S. It will probably take years of bike commuting to make up for my carbon credits, but most meetings I attend don't have viable alternatives in venues like Second Life or World of Warcraft. Anyway, as I sit writing on an airplane, I think I could revise the title for Dr. Seuss' famous book to "Oh the…
One of the interesting things I learned today was that many people are calling for the genome sequences of the chimps and Macaques to be finished.
This is especially amusing because the human genome isn't quite done. We're primates, too! Why not finish our genome?
[I blame these new-found revelations on Twitter. Despite my youngest daughter's warning that only old people use Twitter, I've joined my SciBlings and taken the plunge. (you can even follow me! @digitalbio).
Now, I get to indulge my geeky tendencies while waiting in line at the grocery store. I just type #cshl and voila! I…
We always enjoy home science experiments and it was fun the other night to learn about a new experiment we could try with our teenage daughter and an iPhone.
As it turned out, the joke was on us.
My husband is an enthusiastic fan of the iPhone store. Last night, he downloaded this application called "Army Knife."
This application has, I kid you not, the following nine items:
unit converter - these are always helpful, especially if you travel
ultrasonic whistle
protractor
Heart (beats per minute) counter
measuring tape
digital caliper
Two levels
flashlight
emergency SOS light
Some of…
Warfarin, a commonly used anti-clotting drug, sold under the brand name of Coumadin, has a been a poster child for the promise of pharmacogenomics and personalized medicine.
The excitement has come from the idea that knowing a patient's genotype, in this case for the VKORC1 and CYP2C9 genes, would allow physicians to tailor the dose of the drug and get patients the correct dose more quickly.
And it seems obvious that a test that would allow doctors to predict your ability to metabolize warfarin, would be a great thing, right?
Figure 1. Human Cytochrome P450 Cyp2c9 bound to Warfarin…
Nick's post on Amantadine resistance in swine flu was so interesting, I had to look at the protein structures myself.
I couldn't find any structures with the S31N mutation that Nick discussed, but I did find some structures with the M2 protein and Amantadine. Not only are these structures beautiful, but you can look at them and see how the protein works and how the drug prevents the protein from functioning.
As Nick mentions, the M2 protein from influenza makes a channel for hydrogen ions within the viral membrane. The channel controls the pH inside the virus by opening and closing.…
No more delays! BLAST away!
Time to blast. Let's see what it means for sequences to be similar.
First, we'll plan our experiment. When I think about digital biology experiments, I organize the steps in the following way:
A. Defining the question
B. Making the data sets
C. Analyzing the data sets
D. Interpreting the results
I'm going intersperse my results with a few instructions so you can repeat the things that I've done…
We'll have a blast, I promise! But there's one little thing we need to discuss first...
I want to explain why I'm going to use nucleotide sequences for the blast search. (I used protein the other day). It's not just because someone told me too, there is a solid rational reason for this.
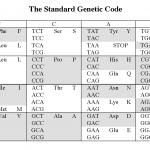
The reason is the redundancy in the genetic code.
Okay, that probably didn't make any sense to those of you who didn't already know the answer. Here it is.
The picture above shows the human genetic code (there are at least 16 variations on this, but that's another story). Each middle cell in the…
We had a great discussion in the comments yesterday after I published my NJ trees from some of the flu sequences.
If I list all the wonderful pieces of advice that readers shared, I wouldn't have any time to do the searches, but there are a few that I want to mention before getting down to work and posting my BLAST results.
Here were some of the great suggestions and pieces of advice;
1. Do a BLAST search. Right! I can't believe I didn't do that first thing, I think the trees I got surprised me so much all sense flew out of my brain.
2. Show us the multiple alignments. Okay. I'll…
We were joking about this a couple of hours ago, but I just picked up the phone and learned that two middle schools will be closed for the week.
Aki Kurose and Stevens middle schools are closed from tomorrow to May 8th. Yikes!
Last night, the phone rang at 9:22 pm. I quickly glanced at the caller ID. Hmmm. Why is the Seattle School district calling us at this time of night?
Apparently the swine flu has come to Seattle and the school district thought we should know.
Those messages are helpful if you're a parent, but they don't tell much about the rest of the world.
Health Map is a really wonderful, user-friendly, resource for following the epidemic.
When you get to Health Map , choose Select None to clear the map.
Then select Swine Flu.
You'll see a Google map with markers representing reports. The colors show…
I'm teaching an on-line bioinformatics course this semester for Austin Community College. They are in Texas of course, but I am in Seattle. This presents a few interesting challenges and some minor moments of amusement.
Today, the school sent all the faculty emails telling us to stay home if we're sick.
Got it. If I think I have flu, I will not fly to Texas.
Instead, I'll stay home and watch videos on coughing without contaminating others.
Watch "Why don't we do it in our sleeves?" and find out how you rank on the safe coughing scale.
What tells us that this new form of H1N1 is swine flu and not regular old human flu or avian flu?
If we had a lab, we might use antibodies, but when you're a digital biologist, you use a computer.
Activity 4. Picking influenza sequences and comparing them with phylogenetic trees
We can get the genome sequences, piece by piece, as I described in earlier, but the NCBI has other tools that are useful, too.
The Influenza Virus Resource will let us pick sequences, align them, and make trees so we can quickly compare the sequences to each other.
This is how I got the sequences that I wrote about…
This afternoon, I was working on educational activities and suddenly realized that the H1N1 strain that caused the California outbreak might be the same strain that caused an outbreak in 2007 at an Ohio country fair.
UPDATE: I'm not so certain anymore that the strains are the same. I'm doing some work with nucleic acid sequences to look further at similarity.
Here's the data.
Once I realized that the genome sequences from the H1N1 swine flu were in the NCBI's virus genome resources database, I had to take a look.
And, like eating potato chips, making phylogenetic trees is a little bit…
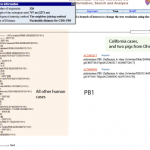
I was pretty impressed to find the swine flu genome sequences, from the cases in California and Texas, already for viewing at the NCBI.
You can get them and work them, too. It's pretty easy. Tomorrow, we'll align sequences and make trees.
Activity 3: Getting the swine flu sequence data
1. Go to the NCBI, find the Influenza Virus Resource page and follow the link to:
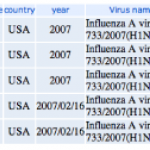
04/27/2009: Newest swine influenza A (H1N1) sequences.
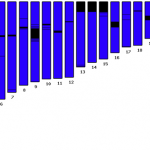
2. You'll see a page that looks like this:
Each column heading is a name of a segment of the influenza genome. You can see there are eight of these. Each segment…
I'm a big of learning from data. There are many things we can learn about swine flu and other kinds of flu by using public databases.
In digital biology activity 1, we learned about the kinds of creatures that can get flu. Personally, I'm a little skeptical about the blowfly, but...
Now, you might wonder, what kinds of flu do these different creatures get? Are they all getting H1N1, or do they get different variations? What are H and N anyway?
We can discuss all of these, but for now, lets see what kinds of flu strains infect different kinds of creatures.
Activity 2. What flu infects…
Genome sequences from California and Texas isolates of the H1N1 swine flu are already available for exploration at the NCBI. Let's do a bit of digital biology and see what we can learn.
Activity 1. What kinds of animals get the flu?
For the past few years we've been worrying about avian (bird). Now, we're hearing about swine (pig) flu.
All of this news might you wonder just who gets the flu besides pigs, birds, and humans. We can find out by looking at the data.
Over the past few years, researchers have been sequencing influenza genomes and depositing those genomes in public…
Usually, I'm kind of a hermit, but I heard there would be Bacon and so, I'm going to leave the house next Weds, night and attend a science blogging event at the University of Washington.
I think it will be fun. Not only will there be Bacon, there will be pizza and other bloggers, who may or may not be entertaining.
Others who will be there (besides me) are:
Dave Bacon, Quantom Pontiff
Maria Brumm, Green Gabbro, who is NOT really an intern with the Discovery Institute
Keith Seinfeld, KPLU, Science and Wonder
http://kplu.wordpress.com/
Alan Boyle, MSNBC, Cosmic Log
Eric Steig, UW Earth…
A couple of years ago, I answered a reader's question about the cost of genome sequencing. One of my readers had asked why the cost of sequencing a human genome was so high. At that time, I used some of the prices advertised by core labs on the web and the reported coverage to estimate the cost of sequencing Craig Venter's genome. As you can imagine, the cost of sequencing has dropped quite a bit since then.
In 2007, Genome Technology reported the cost of sequencing Venter's genome was $70 million. Watson's genome at only $2 million, was a bargain.
Why was Watson's genome so cheap?
Even…