tags: researchblogging.org, evolution, dinosaurs, birds, Tyrannosaurus rex, ornithology, paleontology
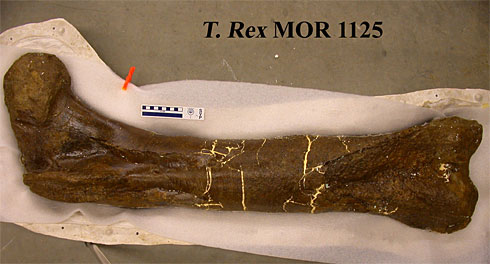
The Tyrannosaurus rex femur from which researcher Mary Higby Schweitzer of North Carolina State University recovered soft tissue.
Image: Science.
It wasn't too long ago that paleontologists thought that fossilization was a process where all biological material was replaced with inert stone. However, in 2005, Mary Higby Schweitzer of North Carolina State University rocked the paleontological world when she recovered a still-elastic blood vessel from inside a fractured thigh bone fossil of a Tyrannosaurus rex that lived 68 million years ago. Recent phylogenetic analyses of this isolated tissue by a team of scientists reveals that the closest living relative of T. rex is none other than .. the humble chicken.
But you already knew that, right? Well, I suspect the reason that this paper was published in Science, even though it reports old news, is because it is truly amazing that scientists are able to obtain useful evolutionary information -- phylogenetic signal -- from a 68-million-year-old protein, and thus, this finding has the potential to revolutionize the world of paleontology.
"I'd call it a milestone," says paleontologist Hans Larsson of McGill University in Montreal. "Dinosaurs will enter the field of molecular biology and really slingshot paleontology into the modern world."
This T. rex fossil was unearthed in Hell Creek, Montana, in 2003 by paleontologist Jack Horner, where it was buried under 1,000 cubic yards of rock. The helicopter used to remove the fossils from the site was unable to carry the intact femur back to the lab, so Horner broke the fossil into pieces and in doing so, discovered small pieces of elastic tissue within.
"For centuries it was believed that the process of fossilization destroyed any original material, consequently no one looked carefully at really old bones," reports Schweitzer. "Finding any soft tissues in dinosaur bones greatly surprised us."
Her team isolated and purified the soft tissues from the fossilized femur and examined them under a microscope. These tissues were similar in appearance to the cells and blood vessels found in ostrich bones. In fact, this purified material looked a lot like collagen -- a class of connective tissues found within all animals.
Several biochemical tests confirmed this. To determine if these collagen molecules were contaminants or they truly were from the T. rex fossil, Schweitzer teamed up with her colleague, John Asara at Harvard Medical School and Beth Israel Deaconess Medical Center who is an expert in mass spectrometry. Mass spectrometry is used to identify minute amounts of proteins and other molecules by measuring their atomic properties. Previously, Asara and Schweitzer collaborated to identify proteins from mammoth remains that were between 160,000 to 400,000 years old. Asara was eager to find out if mass spectrometry could be used to reveal useful information from older molecules, such as these soft tissues recovered from T. rex.
Collagen is a highly conserved protein, meaning that its chemical structure has not changed much from one animal species to another throughout the millenia. Thus, it was highly likely that the sequence of these putative dinosaur collagen proteins would be identical to other species. But to gain useful evolutionary information, these amino acid sequences must be unique because it is through sequence differences that ancestry is revealed.
Analyzing the protein was challenging. After purifying the collagen, researchers had less than a billionth of a gram of the material for mass spectrometry analysis. However, Asara successfully decoded the amino acid sequences from seven pieces of these collagen molecules. When these peptide sequences were entered into the genetic database using BLAST, Asara's team made a startling discovery: several of their newly identified sequences did not specifically match any one species.
"Out of seven total sequences, we had three that matched chicken uniquely," said Asara. "We had another that matched frog uniquely, one that matched newt uniquely, and a couple that matched multiple sequences."
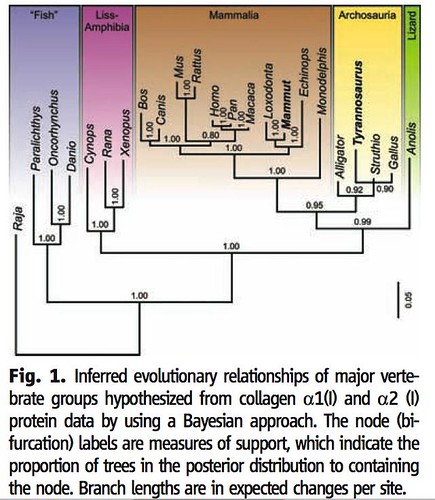
The team used four different statistical methods to compare these peptide sequences to 21 modern animals (including more than 70 sequences from the mammoth proteins that Asara and Schweitzer had previously analyzed) and to analyze the differences to infer their evolutionary relationships. Three of these four analyses inferred the evolutionary relationships between T. rex and modern animals in the same way;
As you can see in figure 1, these analyses strongly suggest that these new collagen molecules did indeed originate from the T. rex fossil because it places T. rex close to both modern ostriches and chickens and somewhat more distant from modern alligators in the group Archosauria (yellow box). This analysis also reveals that such proteins, when they are found in the future, can be used to infer evolutionary relationships between modern and ancient animals. Further, this study provides the exciting opportunity for paleontologists peek into the past, to test their understanding of dinosaur evolution, which is based solely on meticulous examination and analyses of fossilized bones: will peptide sequences confirm or conflict with their current paradigm?
"Most people believe that birds evolved from dinosaurs, but that's all based on the architecture of the bones," mentioned Asara. "This allows you to get the chance to say, 'Wait, they really are related because their sequences are related.' We didn't get enough sequences to definitively say that, but what sequences we got support that idea."
It is not yet known how many other fossils will present researchers with the opportunity to analyze more proteins as these T. rex and mammoth proteins were analyzed.
"Nature has to give you the opportunity to do this first," Asara said philosophically.
But already, Schweitzer's team is planning to go on a worldwide tour of dinosaur sites, looking for more fossil bones to crack open, in search of organic molecules to analyze.
Source
Organ, C.L., Schweitzer, M.H., Zheng, W., Freimark, L.M., Cantley, L.C., Asara, J.M. (2008). Molecular Phylogenetics of Mastodon and Tyrannosaurus rex. Science, 320(5875), 499-499. DOI: 10.1126/science.1154284
BBCNews (quotes).

It used to be, we'd argue over roasted chicken legs;
Now, I keep a wary eye when eating scrambled eggs.
Cool! I'm waiting to see what they'll get from the mummified dinosaur. In the mean time, I think I'll watch my back next time I walk into a KFC.
p.s. ROFL!!!! Cuttlefish, you've just won the Ogden Nash Award for Short, Pithy Verse!
Does this now mean that human tastes like T. rex? Mmm...
A molecular phylogeny including a dinosaur... my gosh I never thought I'd see this! It makes me giddy with excitement just looking at it.
Think this will finally put a nail in the coffin of the thecodont-ancestry school of thought regarding bird origins? Unless you consider tyrannosaurs, etc, to be derived birds...
~ Nick
So you have a bone to pick with them or what?
Just kidding. Nice analysis.
I just finished reading this as opposed to skimming thru it which is what I did before, and I have to wonder, about your last sentence, why "Schweitzer's team is planning to go on a worldwide tour of dinosaur sites," instead of asking museums to look over their collection for candidates. Or is that what you mean by "sites"? Museums would be immediate (relatively speaking!), and a lot cheaper.
Which brings me to my next question: is there any way to tell from some sort of examination or testing of bones, or from characteristics of the location from which the bone came, which might be good candidates - or which might be better candidates than others - for this purpose?
Some kind of right-tuned MRI should work...or not?