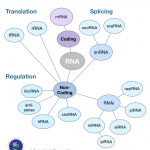
bioinformatics
It's time for the annual blog about the annual Nucleic Acids Research (NAR) database issue. This is the 24th database issue for NAR and the seventh blog for @finchtalk. Like most years I have no idea what I'm going to write about until I start reading the new issue. Something always inspires me.
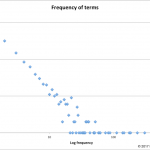
This year's inspiration came from missing data.
In 2017, NAR lists 1662 databases or 23 fewer than last year.
As summarized in the database issue's introduction, Galperin, Fernández-Suarez, and Rigden tell us this year's issue has 152 papers. 54 of those describe new databases, 98…
Computers, biological data (molecular sequences, structures, and other data), websites, and databases are integral to modern research. Innovations like precision, or personalized medicine, expect a certain level of patient participation, and our future food and environmental sustainability will require that society can access a multitude of computer-based resources. Thus, higher education has an important role in providing students with employable skills as well as the ability to use data to make important personal and societal decisions. Toward that goal it is worthwhile understanding…
Pull a spaghetti noodle out of a box of pasta and take a look. It's long and stiff. Try to bend it and it breaks. But fresh pasta is pliable. It can fold just like cooked noodles.
When students first look at an amino acid sequence, a long string of confusing letters, they often think those letters are part of a chain like an uncooked spaghetti noodle. Stiff and unbending, with one end far from the other.
Molecular modeling apps let us demonstrate that proteins are a bit more like fresh pasta.
If we apply rainbow colors (Red Orange Yellow Blue…
Sometimes when you go digging through the databases, you find unexpected things.
When I was researching the previous posts on insulin structure and insulin evolution, I found something curious indeed.
Human insulin, colored by rainbow. Image from the Molecule World iPad app by Digital World Biology.
I wanted to find out how many different organisms made insulin, so I used a database at the NCBI called Blink. Blink is a database of protein blast search results. Using Blink can save you lots of time because…
On pinene and inhibiting enzymes.
People of a certain age may remember a series of really funny commercials featuring Euell Gibbons and his famous question about whether you've ever eaten a pine tree. "Some parts are edible" said Euell.
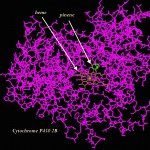
Perhaps some parts are, but other pine tree products aren't so nourishing. Crystallography365, aka @Crystal_in_city had a couple of fun blog posts about pinene, a chemical made by pine trees, that also inhibits cytochrome P450 2B6.
I was inspired by their posts and by my experience with Cytochrome P450 to go a little…
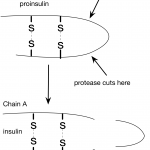
In my last post, I wrote about insulin and interesting features of the insulin structure. Some of the things I learned were really surprising. For example, I was surprised to learn how similar pig and human insulin are. I hadn't considered this before, but this made me wonder about the human insulin we used to give to one of our cats. How do cat and human insulin compare?
It turns out, that all vertebrates produce insulin, even frogs and zebra fish. Human preproinsulin is only 110 amino acids long and even human and fish insulin are pretty similar…
What’s the first you think about when you see a spider? Running away? Danger? Fairies? Spiderman?
Do you wonder if spider silk is really strong enough to stop a train, like they showed in Spiderman 2?
Whatever your thoughts, you’re probably not thinking about 3D printing in space. Yet, the time might be near when astronauts will be using 3D printers filled with spider silk to make replacement parts in space. I learned about this idea in a presentation from Dr. Ron Sims, Utah State University, at the Bioman conference at Salt Lake Community College a couple of…
A few weeks back, we published a review about the development and role of the human reference genome. A key point of the reference genome is that it is not a single sequence. Instead it is an assembly of consensus sequences that are designed to deal with variation in the human population and uncertainty in the data. The reference is a map and like a geographical maps evolves though increased understanding over time.
From the Wiley On Line site:
Abstract
Genome maps, like geographical maps, need to be interpreted carefully. Although maps are essential to exploration and navigation they…
In our series on why $1000 genomes cost $2000, I raised the issue that the $1000 genome is a value based on simplistic calculations that do not account for the costs of confirming the results. Next, I discussed how errors are a natural occurrence of the many processing steps required to sequence DNA and why results need to be verified. In this and follow-on posts, I will discuss the four ways (oversampling, technical replicates, biological replicates, and cross-platform replicates) that results can be verified as recommended by Robasky et. al. [1].
The game Telephone teaches us how a…
By @finchtalk (Todd Smith)
In 2014 and beyond Finchtalk will be contributing to Digitalbio’s blog at this site. We kick off 2014 with Finchtalk’s traditional post on the annual database issue from Nucleic Acids Research (NAR).
Biological data and databases are ever expanding. This year was no exception as the number of databases tracked by NAR grew from 1512 to 1552. In the leadoff introduction [1] the authors summarize this year’s issue and the status of the NAR index. The 21st issue includes 185 articles with 58 new databases and 123 updates. In the 1552 database repository, 193 had their…
I found this post titled "What scientists really want from digital publishing", and, after reading it, I'm pretty certain this scientist doesn't want what's offered. Before I get to the details, here's what computer scientst Philip Bourne offers:
"as a scientist I want an interaction with a publisher that does not begin when the scientific process ends but begins at the beginning of the scientific process itself"
I actually want to do away with publishers--I see them as a necessary evil, including the Open Access ones. What I really want is to be able to communicate to other scientists (and…
I got to spend last week in sunny California. I forgot how wonderful it is to sit and eat lunch outside! I was participating in a workshop held at the Department of Energy's Joint Genome Institute (JGI). The workshop was entitled Microbial Genomics and Metagenomics. Basically I spent the week learning about different tools that are available to help biologists deal with the data flood that has come out (and continues to flow faster and faster) of sequencing technologies that continue to get faster and cheaper.
Since microbes are not exactly easy to observe with ones eyes,…
I had the good fortune on Thursday to hear a fascinating talk on deep transcriptome analysis by Chris Mason, Assistant Professor, at the Institute for Computational Biomedicine at
Cornell University.
Several intriguing observations were presented during the talk. I'll present the key points first and then discuss the data.
These data concern the human transcriptome, and at least some of the results are supported by follow on studies with data from the pigmy tailed macaque.
Some of the most interesting points from Mason's talk were:
A large fraction of the existing genome…
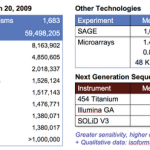
Gabe Rudy, blogging at our 2 snps, has a really good introduction to sequencing technology and its history. It's worth the read, but I don't entirely agree with the reason given for why ABI SOLiD lost out to Illumina:
Coming to market at the same time, but seeming to have just missed the wave, was the Applied Biosystems (ABI) SOLiD system of parallel sequencing by stepwise ligation. Similar to the Solexa technology of creating extremely high throughput short reads cheaply, SOLiD has the added advantage of reading two bases at a time with a florescent label. Because a single base pair…
One of my hobbies lately has been to get either RNA seq or microarray data from GEO and do quick analyses. Not only is this fun, I can find good examples to use for teaching biology.
One of these fun examples comes from some Arabidopsis data. In this experiment, some poor little seedlings were taken out of their happy semi-liquid culture tubes and allowed to dry out. This simulated drought situation isn't exactly dust bowls and hollow-eyed farmers, but the plants don't know that and most likely respond in a similar way.
We can get a quick idea of how the plants feel about their situation…
Plega sp. (Mantispidae)
Who was the source of Monday's DNA? As many of you discerned from the online Genbank database, the sequence came from Plega dactylota, a Neuropteran insect in the family Mantispidae.
10 points to Aaron Hardin, who guessed it first.
For future reference, these genetic puzzles are only slightly more complicated than a Google search. Go to NCBI's BLAST page, select "nucleotide blast" (because we have nucleotide data), click the box for "others" to get you out of the human genome, enter the sequence in the search box, and click the "BLAST" button. Any significant…
Is there a place for citizen scientists in the world of digital biology?
Many of the citizen science projects that I've been reading about have a common structure. There's a University lab at the top, outreach educators in the middle, and a group of citizens out in the field collecting data.
After the data are collected, they end up in a database somewhere and the University researchers analyze them and write papers. At least that's my impression so far.
It seems to me, that with all kinds of databases out there, on-line, there should be plenty of opportunity for both citizens and student…
These days, DNA sequencing happens in one of three ways.
In the early days of DNA sequencing (like the 80's), labs prepared their own samples, sequenced those samples, and analyzed their results. Some labs still do this.
Then, in the 90's, genome centers came along. Genome centers are like giant factories that manufacture sequence data. They have buildings, dedicated staff, and professional bioinformaticians who write programs and work with other factory members to get the data entered, analyzed, and shipped out to the databases. (You can learn more about this and go on a virtual tour in this…
One of the exciting things about bacterial genomics in that, within a year, we'll definitely be in the era of the $1,500 bacterial genome, although that's probably an overestimate. This cost includes everything: labor, sequencing, genome assembly, and genome annotation. While sequencing is highly automated, and has been turned into a production process, akin to a factory, high quality genome annotation, until very recently, has not. Automated software gets about 95% of gene calls right, but the other five percent differs based on the algorithm used.
Unfortunately, this means that, instead…
ScienceBlogling Sandra Porter asks a bunch of questions, including:
What do you call a biologist who uses bioinformatics tools to do research, but doesn't program?
My snarky answer is: a biologist.
Since I work at a genome center where I'm up to my eyeballs in computational biologists, bioinformaticists, and computational engineers (this is a good thing), I was going to write a lengthy response, but then I read one of the commenters, who very pithily stated the differences (boldface mine):
computational biology (the specialists, focusing on developing and applying theoretical biology)…