Genetics & Molecular Biology
It's well understood in science education that students are more engaged when they work on problems that matter. Right now, Zika virus matters. Zika is a very scary problem that matters a great deal to anyone who might want to start a family and greatly concerns my students.
I teach a bioinformatics course where students use computational tools to research biology. Since my students are learning how to use tools that can be applied to this problem, I decided to have them apply their new bioinformatics skills to identify drugs that work against Zika…
"By night all cats are gray" - Miguel Cervantes in Don Quixote
I've always liked Siamese cats. Students do, too. "Why Siamese cats wear masks" is always a favorite story in genetics class. So, when I opened my January copy of The Science Teacher, I was thrilled to see an article on Siamese cat colors and proteins AND molecular genetics (1).
In the article, the authors (Todd and Kenyon) provide some background information on the enzymatic activity of tyrosinase and compare it to the catechol oxidase that causes fruit to brown, especially apples.…
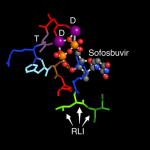
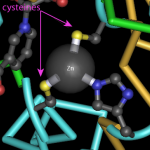
We've been fans of the Molecule of the Month series by David Goodsell, for many years. Not only is Dr. Goodsell a talented artist but he writes very clear descriptions of the ways molecules like proteins, RNA, and DNA work together and function inside a cell.
To learn about proteins and their activities, I like to go directly to the Molecule of the Month page, where I can find a list of articles organized by molecule type and name. Many of these articles can also be downloaded in a PDF format.
A really nice of his articles is that he includes PDB IDs for all the…
A few weeks back, we published a review about the development and role of the human reference genome. A key point of the reference genome is that it is not a single sequence. Instead it is an assembly of consensus sequences that are designed to deal with variation in the human population and uncertainty in the data. The reference is a map and like a geographical maps evolves though increased understanding over time.
From the Wiley On Line site:
Abstract
Genome maps, like geographical maps, need to be interpreted carefully. Although maps are essential to exploration and navigation they…
A key concept in science is molecular scale. DNA is a fascinating molecule in this regard.
While we cannot "see" DNA molecules without the aid of advanced technology, a full length DNA molecule can be very long. In human cells, other than sperm and eggs, six billion base pairs of DNA are packaged into 22 pairs of chromosomes, plus two sex chromosomes. Each base pair is 34 angstroms in length (.34 nanometers, or ~0.3 billionths of a meter), so six billion base pairs (all chromosomes laid out head to toe) form a chain that's two-meters long. If we could hang this DNA chain from a hook, it…
Replication fork - http://en.wikipedia.org/wiki/Telomere.
Organisms with linear chromosomes have to solve the problem that DNA replication makes them shorter. This is due to the fact that DNA polymerase can only add bases to the terminal 3'-OH of a DNA chain. The DNA replication initiation complex uses RNA primers to provide the initial 3'-OH and to initiate "lagging" strand synthesis. While one strand can be copied all the way to the end of a chromosome, the other, lagging strand, must be primed at short intervals in order to provide a 3' OH group for DNA polymerase as the…
Getting an accurate genome sequence requires that you collect the data at least twice argue Robasky, Lewis, and Church in their recent opinion piece in Nat. Rev. Genetics [1].
The DNA sequencing world kicked off 2014 with an audacious start. Andrew Pollack ran an article in the New York Times implying that 100,000 genomes will be the new norm in human genome sequencing projects [2]. The article focused on a collaboration between Regeneron and Geisinger Health in which they plan to sequence the exomes (the ~2% of the genome that encodes proteins and some non-coding RNA) of 100,000…
For the past few days I've been avidly following Daniel MacArthur's tweets from the Personal Genome Conference at Cold Spring Harbor(@dgmacarthur #cshlpg).
The Personal Genomics tweets aren't just interesting because of the science, they're interesting because MacArthur and others have started to take on the conventional dogma in genetic ethics.
For years, there has been a strong message from the clinical genetics and genetics education community that genetic information is dangerous.
Unlike the other medical tests we're continually urged to get (mammograms, blood pressure readings, sugar…
This morning I attended a "bloggers-only" conference call with Dr. Eric Green and the folks from the NIH Human Genome Research Institute (NHGRI) to hear about NHGRI's new strategic plan. The new plan represents a shift away from viewing the genome through a lens marked "for research use only" and towards the goal of making the genome useful as a clinical tool. As a consequence, we will see a greater emphasis on funding activities that support clinical work. For example, it's not always clear how variations in the genome are related to disease. NHGRI might fund projects that help sort and…
One of my hobbies lately has been to get either RNA seq or microarray data from GEO and do quick analyses. Not only is this fun, I can find good examples to use for teaching biology.
One of these fun examples comes from some Arabidopsis data. In this experiment, some poor little seedlings were taken out of their happy semi-liquid culture tubes and allowed to dry out. This simulated drought situation isn't exactly dust bowls and hollow-eyed farmers, but the plants don't know that and most likely respond in a similar way.
We can get a quick idea of how the plants feel about their situation…
You might think the coolest thing about the Next Generation DNA Sequencing technologies is that we can use them to sequence long-dead mammoths, entire populations of microbes, or bits of bone from Neanderthals.
But you would be wrong.
Sure, those are all cool things to do, but Next Generation DNA sequencing (or NGS for short) can give us answers to questions that are far, far more interesting.
With NGS, we can look at entire transcriptomes (!!) together with the proteins that make them and the DNA modifications that help regulate them. If we compare a cell to music, a genome sequence…
No more delays! BLAST away!
Time to blast. Let's see what it means for sequences to be similar.
First, we'll plan our experiment. When I think about digital biology experiments, I organize the steps in the following way:
A. Defining the question
B. Making the data sets
C. Analyzing the data sets
D. Interpreting the results
I'm going intersperse my results with a few instructions so you can repeat the things that I've done…
We had a great discussion in the comments yesterday after I published my NJ trees from some of the flu sequences.
If I list all the wonderful pieces of advice that readers shared, I wouldn't have any time to do the searches, but there are a few that I want to mention before getting down to work and posting my BLAST results.
Here were some of the great suggestions and pieces of advice;
1. Do a BLAST search. Right! I can't believe I didn't do that first thing, I think the trees I got surprised me so much all sense flew out of my brain.
2. Show us the multiple alignments. Okay. I'll…
What tells us that this new form of H1N1 is swine flu and not regular old human flu or avian flu?
If we had a lab, we might use antibodies, but when you're a digital biologist, you use a computer.
Activity 4. Picking influenza sequences and comparing them with phylogenetic trees
We can get the genome sequences, piece by piece, as I described in earlier, but the NCBI has other tools that are useful, too.
The Influenza Virus Resource will let us pick sequences, align them, and make trees so we can quickly compare the sequences to each other.
This is how I got the sequences that I wrote about…
This afternoon, I was working on educational activities and suddenly realized that the H1N1 strain that caused the California outbreak might be the same strain that caused an outbreak in 2007 at an Ohio country fair.
UPDATE: I'm not so certain anymore that the strains are the same. I'm doing some work with nucleic acid sequences to look further at similarity.
Here's the data.
Once I realized that the genome sequences from the H1N1 swine flu were in the NCBI's virus genome resources database, I had to take a look.
And, like eating potato chips, making phylogenetic trees is a little bit…
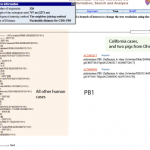
I was pretty impressed to find the swine flu genome sequences, from the cases in California and Texas, already for viewing at the NCBI.
You can get them and work them, too. It's pretty easy. Tomorrow, we'll align sequences and make trees.
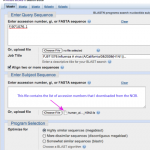
Activity 3: Getting the swine flu sequence data
1. Go to the NCBI, find the Influenza Virus Resource page and follow the link to:
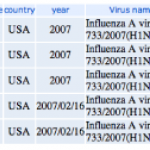
04/27/2009: Newest swine influenza A (H1N1) sequences.
2. You'll see a page that looks like this:
Each column heading is a name of a segment of the influenza genome. You can see there are eight of these. Each segment…
I'm a big of learning from data. There are many things we can learn about swine flu and other kinds of flu by using public databases.
In digital biology activity 1, we learned about the kinds of creatures that can get flu. Personally, I'm a little skeptical about the blowfly, but...
Now, you might wonder, what kinds of flu do these different creatures get? Are they all getting H1N1, or do they get different variations? What are H and N anyway?
We can discuss all of these, but for now, lets see what kinds of flu strains infect different kinds of creatures.
Activity 2. What flu infects…
Genome sequences from California and Texas isolates of the H1N1 swine flu are already available for exploration at the NCBI. Let's do a bit of digital biology and see what we can learn.
Activity 1. What kinds of animals get the flu?
For the past few years we've been worrying about avian (bird). Now, we're hearing about swine (pig) flu.
All of this news might you wonder just who gets the flu besides pigs, birds, and humans. We can find out by looking at the data.
Over the past few years, researchers have been sequencing influenza genomes and depositing those genomes in public…
A couple of years ago, I answered a reader's question about the cost of genome sequencing. One of my readers had asked why the cost of sequencing a human genome was so high. At that time, I used some of the prices advertised by core labs on the web and the reported coverage to estimate the cost of sequencing Craig Venter's genome. As you can imagine, the cost of sequencing has dropped quite a bit since then.
In 2007, Genome Technology reported the cost of sequencing Venter's genome was $70 million. Watson's genome at only $2 million, was a bargain.
Why was Watson's genome so cheap?
Even…
In which we search for Elvis, using blastp, and find out how old we would have to be to see Elvis in a Las Vegas club.
Introduction
Once you're acquainted with proteins, amino acids, and the kinds of bonds that hold proteins together, we can talk about using this information to evaluate the similarity between protein sequences. We can easily imagine that if two protein sequences are identical, then those proteins would have the same kind of activity. But what about proteins that are similar in some regions, and not others, or proteins that only share some of the same amino acids in similar…