One of the key biological questions about antibiotic resistance is to what extent is the spread of resistance due to the evolution of new resistant strains versus the spread of existing resistant strains. With MRSA (methicillin resistant Staphylococcus aureus, it's been thought that epidemic spread of a few resistant genotypes (strains) is responsible for much of the increase in MRSA, as shown by this image of pulsed field gel electrophoresis (PFGE) done on a nationwide collection of MRSA:

(from here)
However, methods such as PFGE lack the genetic resolution to determine if gene transfer is occurring within a clone. In a recent paper that looked at one MLST type ('ST5')--a set of strains that are identical at ~3500 base pairs and whose common ancestor arose roughly 2300 years ago--the authors found that clones, as defined by a set of 156 markers ("bi-allelic polymorphisms"), were very region specific (although not perfectly so):
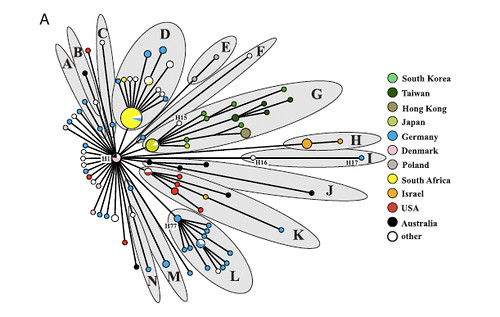
(Minimum spanning tree based on 156 BiPs discovered in 46,483 bps of DNA from each of 135 S. aureus ST5 isolates from a global collection. Circle size is proportional to haplotype frequency, and line length is proportional to the number of mutational steps between haplotypes. (A) Colors indicate the countries of origin of the isolates. Fourteen lineages that consist of at least 2 haplotypes are labeled ''A'' through ''N.'')
The authors also found that the methicillin resistance gene cassette was transferred at least 23 times:
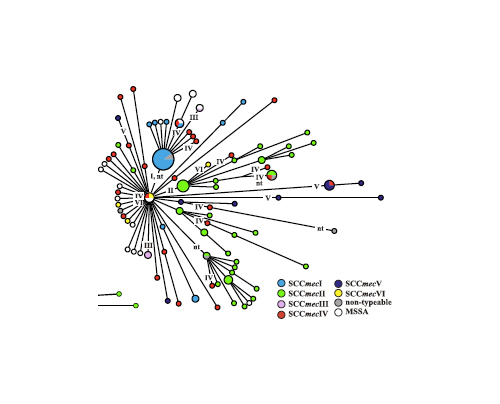
(Colors indicate the type of SCCmecin each of theMRSAisolates. SCCmecelements that are non-typeable on the basis of current PCR schemes probably represent SCCmec variants, because they displayed recombinase gene (ccrB) sequences that were 93% homologous to published ccrB sequences. Roman numerals indicate events of SCCmec acquisition that explain the observed distribution of SCCmec elements in the most parsimonious way. The actual number of SCCmec acquisitions may be higher.)
So what does this mean? At least for this clone--and more work is needed to verify this in other clones (good thing someone is sequencing 40 USA200 MRSA....)--it looks like there isn't pandemic spread of a resistant clone, but, instead repeated gene transfer into a highly prevalent sensitive S. aureus background. In other words, at least in this case, antibiotic resistance appears to evolve and spread locally.

One area often overlooked in the battle against the transfer of dangerous infectious diseases like MRSA in medical environments are the ubiquitous keyboards and mice. Standard keyboards and mice can not be disinfected because germs collect around and in seams and under keys. Recently Man & Machine, Inc. released a White Paper titled, Minimizing Transmission of Infectious Disease in Heath Care Environments by Use of Disinfectable PC Keyboards and Mice. It can be viewed at: http://www.man-machine.com/whitepaper.htm