genomics
You might think the coolest thing about the Next Generation DNA Sequencing technologies is that we can use them to sequence long-dead mammoths, entire populations of microbes, or bits of bone from Neanderthals.
But you would be wrong.
Sure, those are all cool things to do, but Next Generation DNA sequencing (or NGS for short) can give us answers to questions that are far, far more interesting.
With NGS, we can look at entire transcriptomes (!!) together with the proteins that make them and the DNA modifications that help regulate them. If we compare a cell to music, a genome sequence…
A recent paper in Nature Biotechnology reported the first complete human genome to be sequenced using third-generation, single-molecule sequencing technology. The genome sequenced belongs to one Stephen Quake, co-founder of the biotech company Helicos that developed the Heliscope instrument used to perform the analysis. On Genetic Future, ScienceBlogger Dan MacArthur analyzes the paper itself, explaining that while single-molecule sequencing is undoubtedly the direction in which genome analysis is headed, the technology is not yet on par with second-generation sequencing platforms in terms…
...assembly and analysis. The Wellcome Trust has a very good (and mostly accurate) article about the 'next-gen' sequencing technologies. I'm going to focus on bacterial genomics because humans are boring (seriously, compared to two bacteria in the same species, once you've seen one human genome, you've seen them all).
Most of the time, when you read articles about sequencing, they focus on the actual production of raw sequence data (i.e., 'reads'). But that's not the rate-limiting step. That is, we have now reached the point where working with the data we generate is far more time-…
Inspired by this funny satire of mouse genomics research ("Tedious scientists hail uninspiring mouse genome breakthrough"), I dug up this funny article in a newsletter from the genome center at which I work:
30x sequencing of Unicornos
Typically the stuff of legends, the [sequencing center] will sequence DNA from the genus Unicornos to high coverage. Tracing the origin of the unicorn points to a 4th century B.C. Greek doctor named Ctesias who most likely fused details of multiple creatures to create the unicorn from the many tales he heard from Indian travelers in Persia. During that era it…
Blogger Thomas Mailund is an author on a new paper, Ancestral Population Genomics: The Coalescent Hidden Markov Model Approach:
With incomplete lineage sorting (ILS), the genealogy of closely related species differs along their genomes. The amount of ILS depends on population parameters such as the ancestral effective population sizes and the recombination rate, but also on the number of generations between speciation events. We use a hidden Markov model parametrized according to coalescent theory in order to infer the genealogy along a four-species genome alignment of closely related species…
I notice that Fortune has a story on personal genomics up, Genetic sequencing gets personal Biotech firm Illumina will sequence your entire genetic code -- and throw in a Mac -- for $48,000:
So far, personalized genomics make up just a small fraction of Illumina's revenue. High costs keep sequencing out of reach for most people. But prices will fall substantially as the technology improves. In fact experts say costs could reach $1000 within three to five years, making more people privy to their entire genetic code.
...
One area Illumina is not diving into is sequence analysis. Instead, it is…
A few months ago we learned via an unintentionally leaked press release that a team of researchers lead by Nicole Gerardo and Cameron Currie had won a Roche Applied Sciences grant competition. The team will be sequencing the complete genome of 14 players from the ant/fungus/microbe co-evolutionary system, including three attine ants from different genera.
The announcement is now official.
An Acromyrmex queen, with brood, in the fungus garden
There is a reason why there are no dinosaur geneticists - their careers would quickly become as extinct as the 'terrible lizards' themselves. Bones may fossilise, but soft tissues and molecules like DNA do not. Outside of the fictional world of Jurassic Park, dinosaurs have left no genetic traces for eager scientists to study.
Nonetheless, that is exactly what Chris Organ and Scott Edwards from Harvard University have managed to do. And it all started with a simple riddle: which came first, the chicken or the genome?
Like almost all birds, a chicken's genome - its full complement of DNA…
It's never made much sense to me why the pathogenic bacteria Salmonella and Shigella (which is really E. coli) have lost the ability to use lactose (milk sugar). In Shigella, we know that when we restore some lost functions through genetic manipulation (e.g., cadaverine production), they actually prevent these altered Shigella from causing disease. But lactose seems to be a good sugar to be able to grow on--they're exposed to it from time to time (in infants).
The genus Salmonella contains two species: S. enteriditis, which causes disease* and can't use lactose as a carbon source, and S.…
Should any data, not just genomic data, be held hostage by the grant award process?
Hunh? Let me back up...
By way of ScienceBlogling Daniel MacArthur, I came across this excellent post by David Dooling about, among other things, how different genome centers, based on size, have different release policies (seriously, read his post). Dooling writes (boldface mine):
The more interesting question is: why aren't all data and research released rapidly and freely available? Since the Bermuda Principles were agreed to in 1996, all genome sequencing centers have submitted their data, from raw…
I realize that the typical format for blogging is to find something that pisses you off and then rant about it, but I actually like the recent workshop report by NHGRI, "The Future of DNA Sequencing at the National Human Genome Research Institute." (pdf file) While I'll have more to say about the report overall, I liked the section about the Human Microbiome Project (the goal of the HMP is to use sequencing technologies to understand how the microbes that live on us and in us affect health and disease).
I was happy to see that NHGRI still thinks that it has a role in funding the HMP. It's…
...ok, I'll stop. But 99 E. coli commensal genomes will be sequenced. And that's really cool.
I'm always wary of counting chickens before they hatch, but I'm fairly certain at least 99 E. coli genomes will be sequenced (NIAID will continue funding, and more than fifty are already in various stages of sequencing).
And to a considerable extent, it's the Mad Biologist's fault. I'm the crazy bastard who thought of this and proposed it (obviously, many, many others are involved from sequencing to funding, and, last but not least, strain collections).
What makes this project unique is that…
While I'm away at ASM, here's something from the archives for you
When I read Olivia Judson's post about hopeful monsters, I didn't think she used the term correctly (here are some good explanations why), but I was surprised by Jerry Coyne's response.
First, the personal attack on Judson is unwarranted: when we reach the point where the serious challenge to evolutionary biology is the misuse of a discredited decades-old idea, as opposed to the politically powerful anti-science creationist movement, we're in a pretty good place. She made a mistake--I don't think her motives were self-…
Probably not. But genes linked to a high risk of breast cancer? You betcha.
ScienceBlogling Rebecca Skloot has a very good piece about the lawsuit brought by the ACLU against Myriad, the company that owns the patent for the 'breast cancer genes' BRCA1 and BRCA2 (she provides some more background here).
To me, the really galling thing is that Myriad didn't discover these genes, publicly funded research did. The goal of that research is not to enrich patent holders, but to improve human health for society as a whole. The patent drives up diagnosis costs by preventing anyone else from…
One of the interesting things I learned today was that many people are calling for the genome sequences of the chimps and Macaques to be finished.
This is especially amusing because the human genome isn't quite done. We're primates, too! Why not finish our genome?
[I blame these new-found revelations on Twitter. Despite my youngest daughter's warning that only old people use Twitter, I've joined my SciBlings and taken the plunge. (you can even follow me! @digitalbio).
Now, I get to indulge my geeky tendencies while waiting in line at the grocery store. I just type #cshl and voila! I…
Warfarin, a commonly used anti-clotting drug, sold under the brand name of Coumadin, has a been a poster child for the promise of pharmacogenomics and personalized medicine.
The excitement has come from the idea that knowing a patient's genotype, in this case for the VKORC1 and CYP2C9 genes, would allow physicians to tailor the dose of the drug and get patients the correct dose more quickly.
And it seems obvious that a test that would allow doctors to predict your ability to metabolize warfarin, would be a great thing, right?
Figure 1. Human Cytochrome P450 Cyp2c9 bound to Warfarin…
If all has gone well, I'm at Cold Spring Harbor for the Biology of Genomes meeting (actually, I'm here a day early for the Microbiome 'pre-meeting'), so responding to email and comments will be sporadic. You know you're in a fast-moving area when the last figure is assembled Friday morning (because the data were just released), and by Sunday--two days later--your poster is out of date....
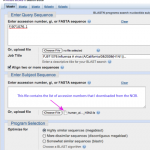
No more delays! BLAST away!
Time to blast. Let's see what it means for sequences to be similar.
First, we'll plan our experiment. When I think about digital biology experiments, I organize the steps in the following way:
A. Defining the question
B. Making the data sets
C. Analyzing the data sets
D. Interpreting the results
I'm going intersperse my results with a few instructions so you can repeat the things that I've done…
We had a great discussion in the comments yesterday after I published my NJ trees from some of the flu sequences.
If I list all the wonderful pieces of advice that readers shared, I wouldn't have any time to do the searches, but there are a few that I want to mention before getting down to work and posting my BLAST results.
Here were some of the great suggestions and pieces of advice;
1. Do a BLAST search. Right! I can't believe I didn't do that first thing, I think the trees I got surprised me so much all sense flew out of my brain.
2. Show us the multiple alignments. Okay. I'll…